

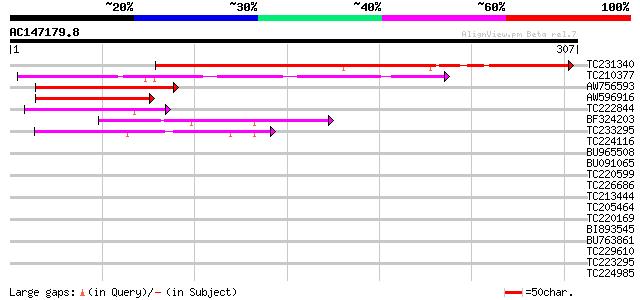
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147179.8 + phase: 0
(307 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC231340 similar to UP|ECR_AEDAE (P49880) Ecdysone receptor (Ecd... 179 1e-45
TC210377 similar to UP|Q8VAG3 (Q8VAG3) Wsv453 (WSSV513), partial... 83 2e-16
AW756593 70 9e-13
AW596916 67 1e-11
TC222844 weakly similar to UP|Q9FFW4 (Q9FFW4) Similarity to heat... 50 1e-06
BF324203 similar to GP|845230|gb|AA maturase {Bensoniella oregon... 48 6e-06
TC233295 similar to UP|Q7KVQ0 (Q7KVQ0) CG4038-PA, partial (6%) 47 8e-06
TC224116 40 0.001
BU965508 40 0.001
BU091065 37 0.011
TC220599 similar to UP|Q9M9E9 (Q9M9E9) F3F9.17 (At1g78290/F3F9_1... 37 0.011
TC226686 homologue to UP|O22512 (O22512) Grr1, complete 35 0.033
TC213444 33 0.12
TC205464 weakly similar to GB|AAP42752.1|30984578|BT008739 At1g5... 32 0.47
TC220169 31 0.62
BI893545 30 1.1
BU763861 30 1.4
TC229610 weakly similar to UP|O22512 (O22512) Grr1, partial (52%) 30 1.4
TC223295 30 1.8
TC224985 homologue to UP|Q93YI3 (Q93YI3) Calcium dependent calmo... 30 1.8
>TC231340 similar to UP|ECR_AEDAE (P49880) Ecdysone receptor (Ecdysteroid
receptor) (20-hydroxy-ecdysone receptor) (20E receptor),
partial (4%)
Length = 971
Score = 179 bits (455), Expect = 1e-45
Identities = 111/232 (47%), Positives = 142/232 (60%), Gaps = 6/232 (2%)
Frame = +2
Query: 80 LRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQHFSSCFFSCHTL 139
LRD S L +DF G ++P+LLK++V YAVSHNV +L + CD + C FSC TL
Sbjct: 5 LRDASLALLKLDFERHGCIEPQLLKRIVKYAVSHNVRQLGISVKCDIRDVPQCVFSCQTL 184
Query: 140 TSLDLCVGP-PRTDERLSFPKSLNLPALTTLSLESFAFCVG---DDGCAEPFSALNSLKN 195
TSL L V P FPKSLNL ALTTL L+ F FC G DD AEPF A L +
Sbjct: 185 TSLKLSVCPRGYIYGSTLFPKSLNLTALTTLHLQHFTFCKGDDDDDDMAEPFYACRRLCD 364
Query: 196 LMILYCNVLDAESLCISSVTLTNLTIVGDPV--YYSTVELSTPSLFTFDFVCYEGIPVLK 253
L I YC V DA LCISS TL +LT+ D +Y V LSTP+L F F G P +
Sbjct: 365 LTIDYCTVKDARILCISSATLVSLTMRSDQSGDFYKIV-LSTPNLCAFAFT---GAPYQQ 532
Query: 254 LCRSKINLSSVKHVNIEVRMWTDYADASLVLLNWLAELANIKSLTLNHTALK 305
L S NLSS++ V+I+ +W+ ++ L+LL+WL ELAN+KSLT++ + L+
Sbjct: 533 LLGS--NLSSLERVSIDAEIWSTSLESPLILLSWLLELANVKSLTVSASTLQ 682
>TC210377 similar to UP|Q8VAG3 (Q8VAG3) Wsv453 (WSSV513), partial (10%)
Length = 1077
Score = 82.8 bits (203), Expect = 2e-16
Identities = 79/243 (32%), Positives = 110/243 (44%), Gaps = 9/243 (3%)
Frame = +3
Query: 5 RKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTI-SSRWRHLWKHLPTLILCSSH 63
+KK K DLPNCVLL+I+ ++ +V+TC + S RW+ L K+L +L S
Sbjct: 249 KKKRQRTEKRRGTGDLPNCVLLQIMEFMNTKYSVQTCVLLSKRWKDL*KYLTSLSF--SR 422
Query: 64 FMRIKSFNR--FVSRI---LCLRD-VSTPLHAVDFLCRGVVD-PRLLKKVVTYAVSHNVE 116
F R R F++ + CL + V +P + L R P LL K N +
Sbjct: 423 FQRDNLCGRGYFITNLCLGFCLVEMVPSPFLILISLGRWFAGAPNLLNK-------QNTQ 581
Query: 117 KLRVHACCDFQHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAF 176
+L + A F F C LT L L + PKSL PAL +L L + F
Sbjct: 582 RLAIDADYIPDCFFPLIFCCQFLTFLKLSIYS-------HLPKSLQFPALKSLHLVNVGF 740
Query: 177 CVGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYS-TVELST 235
D CAEPFS NSL + C +S L +LT+V Y++ + LST
Sbjct: 741 TAIDRSCAEPFSTCNSLNTFLSAGC-----KSPLHIYANLHSLTLVNATRYFAHGIVLST 905
Query: 236 PSL 238
P+L
Sbjct: 906 PNL 914
>AW756593
Length = 370
Score = 70.5 bits (171), Expect = 9e-13
Identities = 33/77 (42%), Positives = 49/77 (62%)
Frame = +3
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNRFV 74
D++S+LP+ +LL ++ +D EAV+TC +S RW +LWK L TL+ +S F + N+F+
Sbjct: 63 DKISELPDNILLHMMDFMDTREAVQTCVLSKRWNNLWKRLSTLLFNTSKFESVFKINKFL 242
Query: 75 SRILCLRDVSTPLHAVD 91
R L RD S L VD
Sbjct: 243 CRFLSDRDDSISLLNVD 293
>AW596916
Length = 455
Score = 66.6 bits (161), Expect = 1e-11
Identities = 31/64 (48%), Positives = 42/64 (65%)
Frame = +3
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNRFV 74
DRLSDLP+ VLL I+ + AV+TC +S RW+ LWK L L L SS F + F++F+
Sbjct: 264 DRLSDLPDFVLLHIMKFMSMKHAVQTCVLSKRWKELWKRLTNLALHSSDFADLAHFSKFL 443
Query: 75 SRIL 78
S +L
Sbjct: 444 SWVL 455
>TC222844 weakly similar to UP|Q9FFW4 (Q9FFW4) Similarity to heat shock
transcription factor HSF30, partial (6%)
Length = 746
Score = 50.4 bits (119), Expect = 1e-06
Identities = 30/81 (37%), Positives = 44/81 (54%), Gaps = 2/81 (2%)
Frame = +1
Query: 9 NDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR-- 66
N+ + DR+S LP+ +L +++ +D AV+TC +S RW L K L L S
Sbjct: 478 NEDHRPDRISALPDSLLFHMINFMDTKSAVQTCVLSKRWNDLSKCLTNLTFNSDLPSEGL 657
Query: 67 IKSFNRFVSRILCLRDVSTPL 87
+SF +F S +L RD S PL
Sbjct: 658 DRSFKKFESWVLSSRDDSYPL 720
>BF324203 similar to GP|845230|gb|AA maturase {Bensoniella oregona}, partial
(3%)
Length = 423
Score = 47.8 bits (112), Expect = 6e-06
Identities = 42/140 (30%), Positives = 64/140 (45%), Gaps = 13/140 (9%)
Frame = +3
Query: 49 HLWKHLPTLILCSSHFMRIKSFNRFVSRILCLRDVSTPLHAVDFLCRGV---VDPRLLKK 105
+LW +P L S FMR+ F +FV +L RD S+ + + + GV D LL K
Sbjct: 3 YLWASVPCLNFSSKSFMRLVDFKKFVLWVLNHRD-SSHVKLLVYYRFGVDYATDQGLLNK 179
Query: 106 VVTYAVSHNVEKLRVHACCDFQHFSS---------CFFSCHTLTSLDL-CVGPPRTDERL 155
V+ YA SH VE+++++ +S F+C +L L+L P L
Sbjct: 180 VIEYAASHGVEEIKINLRAKTAGRTSGSPPVEIPFSLFTCQSLKKLELKDCHPTNGSSSL 359
Query: 156 SFPKSLNLPALTTLSLESFA 175
KSL++ L S+ A
Sbjct: 360 LGCKSLDILHLEQFSMHPVA 419
>TC233295 similar to UP|Q7KVQ0 (Q7KVQ0) CG4038-PA, partial (6%)
Length = 557
Score = 47.4 bits (111), Expect = 8e-06
Identities = 39/142 (27%), Positives = 61/142 (42%), Gaps = 11/142 (7%)
Frame = +3
Query: 14 EDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSS------HFMRI 67
+DR+S LP+ +LL+ILSLL +AV T +S RWR LW + L H +
Sbjct: 63 DDRISHLPDVLLLQILSLLPTKQAVITGILSKRWRPLWPAVSVLDFDDESSPEFHHPGGL 242
Query: 68 KSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKL--RVHACCD 125
F FV +L L D A++ +P + + + H + RV
Sbjct: 243 TGFAEFVYSVLLLHDAP----AIERFRLRCANPNYSARDIATWLCHVARRRAERVELSLS 410
Query: 126 FQHFSS---CFFSCHTLTSLDL 144
+ + C F C T++ + L
Sbjct: 411 LSRYVALPRCLFHCDTVSVMKL 476
>TC224116
Length = 520
Score = 40.4 bits (93), Expect = 0.001
Identities = 18/49 (36%), Positives = 29/49 (58%)
Frame = +2
Query: 10 DYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLI 58
D + D +SDLP ++ IL L +AV+T +SS+WR+ W + L+
Sbjct: 272 DVMGPDLISDLPQSIIESILVQLPIRDAVRTSILSSKWRYKWASITQLV 418
>BU965508
Length = 447
Score = 40.4 bits (93), Expect = 0.001
Identities = 18/50 (36%), Positives = 30/50 (60%)
Frame = +2
Query: 9 NDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLI 58
+D + D +SDLP ++ IL L +AV+T +SS+WR+ W + L+
Sbjct: 278 DDAMGPDLISDLPQSIIESILVQLPIRDAVRTSILSSKWRYKWASITRLV 427
>BU091065
Length = 460
Score = 37.0 bits (84), Expect = 0.011
Identities = 15/41 (36%), Positives = 24/41 (57%)
Frame = +3
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLP 55
+R+ DLP V++ IL L + VKT ++ WR++W P
Sbjct: 90 NRIGDLPMNVIVSILQRLPFQDLVKTSVLARAWRYMWSSTP 212
>TC220599 similar to UP|Q9M9E9 (Q9M9E9) F3F9.17 (At1g78290/F3F9_17), partial
(29%)
Length = 737
Score = 37.0 bits (84), Expect = 0.011
Identities = 44/148 (29%), Positives = 64/148 (42%), Gaps = 6/148 (4%)
Frame = -1
Query: 156 SFPKSLNLPALTTLSLESFAFCVGDDGCAEPFSALNSLKNLMILYC--NVLDAESLCISS 213
S SLNLPAL LS S A N+ NLMIL C ++ SLC S
Sbjct: 596 SSSSSLNLPALHILS-NSSPPAAYSMTIAR*VGVSNTSLNLMILGCFKDL*FIISLCTCS 420
Query: 214 VTL----TNLTIVGDPVYYSTVELSTPSLFTFDFVCYEGIPVLKLCRSKINLSSVKHVNI 269
L NL P+++S +TP + I ++ RS + L+S+ N
Sbjct: 419 SILKPLSMNLIAYNSPLHFSLTSFATPKF-------PDPISLIISYRSMVKLASLN*SN- 264
Query: 270 EVRMWTDYADASLVLLNWLAELANIKSL 297
++ +Y +V+L + ELAN+ L
Sbjct: 263 --SLYNNYQQQGVVVLIEVCELANLSIL 186
>TC226686 homologue to UP|O22512 (O22512) Grr1, complete
Length = 2818
Score = 35.4 bits (80), Expect = 0.033
Identities = 36/141 (25%), Positives = 60/141 (42%), Gaps = 14/141 (9%)
Frame = +1
Query: 95 RGVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQHFSSCFF-----SCHTLTSLDLCVGPP 149
RG R + V A++H L+V + D CH L LDLC P
Sbjct: 634 RGCNSDRGVTNVGLKAIAHGCPSLKVCSLWDVATVGDVGLIEIASGCHQLEKLDLCKCPN 813
Query: 150 RTDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAEPFSALNSLKNLMILYCN------- 202
+D+ L + N P L LS+ES +G++G + +L+++ I C+
Sbjct: 814 ISDKTL-IAVAKNCPNLAELSIESCP-NIGNEG-LQAIGKCPNLRSISIKNCSGVGDQGV 984
Query: 203 --VLDAESLCISSVTLTNLTI 221
+L + S ++ V L +L +
Sbjct: 985 AGLLSSASFALTKVKLESLNV 1047
>TC213444
Length = 530
Score = 33.5 bits (75), Expect = 0.12
Identities = 12/25 (48%), Positives = 20/25 (80%)
Frame = +2
Query: 14 EDRLSDLPNCVLLRILSLLDADEAV 38
EDRLSD+P+C++ ILS ++ +A+
Sbjct: 455 EDRLSDMPDCIIHHILSFMETKDAI 529
>TC205464 weakly similar to GB|AAP42752.1|30984578|BT008739 At1g57790
{Arabidopsis thaliana;} , partial (17%)
Length = 756
Score = 31.6 bits (70), Expect = 0.47
Identities = 15/39 (38%), Positives = 22/39 (55%)
Frame = +1
Query: 9 NDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRW 47
N+Y++ SDLP +L ILS L D+ V+ + RW
Sbjct: 157 NEYLELQTWSDLPTELLELILSRLSLDDNVRASVVCKRW 273
>TC220169
Length = 768
Score = 31.2 bits (69), Expect = 0.62
Identities = 19/66 (28%), Positives = 34/66 (50%), Gaps = 5/66 (7%)
Frame = -2
Query: 160 SLNLPALTTLSLESFAFCVGDD----GCAEP-FSALNSLKNLMILYCNVLDAESLCISSV 214
SLNL L+L FC+G + G +P S L+ L+ +Y +L +++ + ++
Sbjct: 521 SLNLMNTNALALTKIKFCIGKER*IFGRKKPELSCLSFLEKRK*IYIGILQQQNMTVGNM 342
Query: 215 TLTNLT 220
T+ LT
Sbjct: 341 TVGKLT 324
>BI893545
Length = 421
Score = 30.4 bits (67), Expect = 1.1
Identities = 15/41 (36%), Positives = 19/41 (45%)
Frame = +1
Query: 156 SFPKSLNLPALTTLSLESFAFCVGDDGCAEPFSALNSLKNL 196
S + LTTL L +F C C +PF+ LKNL
Sbjct: 247 SLAELFGFTTLTTLHLNNFILCYTGTDCLDPFANCVHLKNL 369
>BU763861
Length = 463
Score = 30.0 bits (66), Expect = 1.4
Identities = 17/41 (41%), Positives = 20/41 (48%)
Frame = +3
Query: 162 NLPALTTLSLESFAFCVGDDGCAEPFSALNSLKNLMILYCN 202
N P +T L L +GDDG A S L NL + YCN
Sbjct: 306 NCPKMTELDLYR-CVRIGDDGLAALTSGCKGLTNLNLSYCN 425
>TC229610 weakly similar to UP|O22512 (O22512) Grr1, partial (52%)
Length = 1428
Score = 30.0 bits (66), Expect = 1.4
Identities = 43/182 (23%), Positives = 75/182 (40%), Gaps = 7/182 (3%)
Frame = +2
Query: 93 LCRGVVDPRLLKKVVTYAVSHNVEKLRVHACCDF-----QHFSSCFFSCHTLTSLDLCVG 147
L RG R + + AV+H LR + + + S CH L LDLC
Sbjct: 140 LIRGSNSERGVTNLGLSAVAHGCPSLRSLSLWNVSTIGDEGVSQIAKRCHILEKLDLCHC 319
Query: 148 PPRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAEPFSALNSLKNLMILYCNVLDAE 207
+++ L + P LTTL++ES +G++G L+++ + C ++
Sbjct: 320 SSISNKGL-IAIAEGCPNLTTLTIESCP-NIGNEGLQAIARLCTKLQSISLKDCPLVGDH 493
Query: 208 SLCISSVTLTNLTIVGDPVYYSTVELSTPSL--FTFDFVCYEGIPVLKLCRSKINLSSVK 265
+ + +NL S V+L T + F+ +C+ G + L LS +K
Sbjct: 494 GVSSLLASASNL---------SRVKLQTLKITDFSLAVICHYGKAITNLV-----LSGLK 631
Query: 266 HV 267
+V
Sbjct: 632 NV 637
>TC223295
Length = 778
Score = 29.6 bits (65), Expect = 1.8
Identities = 16/35 (45%), Positives = 19/35 (53%)
Frame = +1
Query: 45 SRWRHLWKHLPTLILCSSHFMRIKSFNRFVSRILC 79
SRW HLP LIL SS + I+ F VS +C
Sbjct: 1 SRWTKPSLHLPNLIL*SSIILNIQFF*YLVSEYIC 105
>TC224985 homologue to UP|Q93YI3 (Q93YI3) Calcium dependent calmodulin
independent protein kinase (Fragment), partial (14%)
Length = 852
Score = 29.6 bits (65), Expect = 1.8
Identities = 21/73 (28%), Positives = 38/73 (51%)
Frame = +1
Query: 216 LTNLTIVGDPVYYSTVELSTPSLFTFDFVCYEGIPVLKLCRSKINLSSVKHVNIEVRMWT 275
L+ I+G + ++++ TF+ + G+P L +K++ S V+ + VR+
Sbjct: 85 LSEEEIIGLKEMFKSMDIDNSGTITFEEL-KAGLPKLG---TKVSESEVRQLMEAVRILX 252
Query: 276 DYADASLVLLNWL 288
DY A LVLL +L
Sbjct: 253 DYTSAFLVLLYYL 291
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.138 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,064,523
Number of Sequences: 63676
Number of extensions: 309343
Number of successful extensions: 2480
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 2469
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2475
length of query: 307
length of database: 12,639,632
effective HSP length: 97
effective length of query: 210
effective length of database: 6,463,060
effective search space: 1357242600
effective search space used: 1357242600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147179.8


