

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
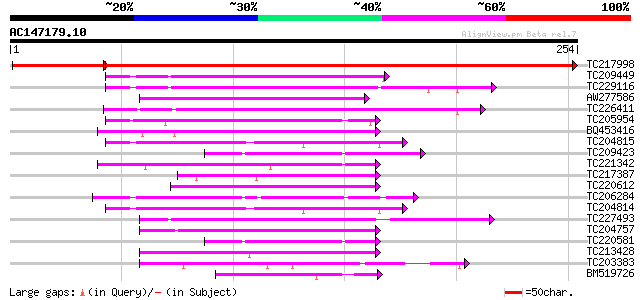
Query= AC147179.10 - phase: 1 /pseudo
(254 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217998 similar to UP|Q8L7U0 (Q8L7U0) AT3g03330/T21P5_25, parti... 377 e-118
TC209449 weakly similar to UP|Q9NYR8 (Q9NYR8) Photoreceptor oute... 69 3e-12
TC229116 similar to UP|PGDH_HUMAN (P15428) 15-hydroxyprostagland... 64 5e-11
AW277586 61 4e-10
TC226411 weakly similar to UP|TRNH_DATST (P50165) Tropinone redu... 59 2e-09
TC205954 similar to UP|Q9FK50 (Q9FK50) Brn1-like protein, partia... 58 5e-09
BQ453416 58 5e-09
TC204815 similar to UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein P... 57 6e-09
TC209423 weakly similar to UP|Q9LUE4 (Q9LUE4) Similarity to oxid... 55 4e-08
TC221342 similar to UP|Q9SY73 (Q9SY73) F14N23.19, partial (74%) 54 5e-08
TC217387 similar to GB|AAO24564.1|27808568|BT003132 At3g06060 {A... 54 7e-08
TC220612 weakly similar to UP|Q9SBM0 (Q9SBM0) Wts2L, partial (66%) 54 7e-08
TC206284 similar to UP|Q8S305 (Q8S305) Short-chain alcohol dehyd... 52 2e-07
TC204814 UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein PM34, complete 52 3e-07
TC227493 similar to UP|FABG_CUPLA (P28643) 3-oxoacyl-[acyl-carri... 52 3e-07
TC204757 similar to UP|Q9ASX2 (Q9ASX2) At1g07440/F22G5_16, parti... 50 1e-06
TC220581 weakly similar to UP|Q9LUE4 (Q9LUE4) Similarity to oxid... 48 5e-06
TC213428 similar to UP|O42866 (O42866) SPAC3G9.02 protein, parti... 48 5e-06
TC203383 similar to UP|Q9FXD7 (Q9FXD7) F12A21.14, partial (21%) 47 6e-06
BM519726 similar to GP|22022575|gb| AT3g03330/T21P5_25 {Arabidop... 47 8e-06
>TC217998 similar to UP|Q8L7U0 (Q8L7U0) AT3g03330/T21P5_25, partial (90%)
Length = 1244
Score = 377 bits (967), Expect(2) = e-118
Identities = 181/211 (85%), Positives = 200/211 (94%)
Frame = +3
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
R V++AES FPDSGVDYM+HNAA+ERPK+S+LDVTEE LKATFDVNV GTITLT+LL P
Sbjct: 399 RIAVEKAESFFPDSGVDYMVHNAAFERPKTSILDVTEEGLKATFDVNVLGTITLTKLLAP 578
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGP 163
FML+RG GHFVVMSSAAGKTPAPGQAVYSASKYALNGYFH+LRSELCQKGIQVTVVCPGP
Sbjct: 579 FMLKRGHGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHTLRSELCQKGIQVTVVCPGP 758
Query: 164 IETANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYW 223
IET+NN+GS+VPSEKRV +E+C ELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYW
Sbjct: 759 IETSNNAGSRVPSEKRVPSERCAELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYW 938
Query: 224 LMDKVGKNRVEAAKEKGNAYSLSLLFGKKKA 254
+MDK+GK+RVEAA++KGN YSLSLL GKKKA
Sbjct: 939 VMDKIGKSRVEAAEQKGNTYSLSLLLGKKKA 1031
Score = 64.7 bits (156), Expect(2) = e-118
Identities = 29/43 (67%), Positives = 32/43 (73%)
Frame = +2
Query: 2 LDYWCQSWNWGNSSSTTCKFRGQVNTICKG*S*PKPSKVTAKR 44
LDYWC WNWG+S T CKFRGQ +CK *S* KPSK TA+R
Sbjct: 206 LDYWC*PWNWGDSG*TVCKFRGQAYYLCKE*S*AKPSKDTAER 334
>TC209449 weakly similar to UP|Q9NYR8 (Q9NYR8) Photoreceptor outer segment
all-trans retinol dehydrogenase, partial (19%)
Length = 1029
Score = 68.6 bits (166), Expect = 3e-12
Identities = 45/129 (34%), Positives = 69/129 (52%), Gaps = 2/129 (1%)
Frame = +1
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
RKVVD + F +D +++NA + + +V +++ TFD NVFG++ + + + P
Sbjct: 268 RKVVDAVVNKF--GRIDVLVNNAGVQCV-GPLAEVPLSAIQNTFDTNVFGSLRMIQAVVP 438
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGP 163
M R +G V + S A P Y+ASK AL+ + +LR EL GI V V PG
Sbjct: 439 HMAVRKQGKIVNVGSVAALASGPWSGTYNASKAALHAFTDTLRLELGHFGIDVVNVVPGA 618
Query: 164 IET--ANNS 170
I + ANN+
Sbjct: 619 ITSNIANNA 645
>TC229116 similar to UP|PGDH_HUMAN (P15428) 15-hydroxyprostaglandin
dehydrogenase [NAD+] (PGDH) , partial (6%)
Length = 1129
Score = 64.3 bits (155), Expect = 5e-11
Identities = 54/201 (26%), Positives = 85/201 (41%), Gaps = 26/201 (12%)
Frame = +2
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
RKVVD + +D +++NA + + + +++ TFD NVFG++ + + + P
Sbjct: 398 RKVVDAVVDKY--GRIDVLVNNAGVQCV-GPLAEAPLSAIQNTFDTNVFGSLRMVQAVVP 568
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGP 163
M + KG V + S A P Y+ASK A + +LR EL GI V + PG
Sbjct: 569 HMATKKKGKIVNIGSVAALASGPWSGAYTASKAAXHALTDTLRLELGHFGIDVVNIVPGA 748
Query: 164 IETANNSGSQVPSEKRVSAEKCV-----------------------ELTIIAATHGLKE- 199
I+ +N S + S R+ K E I LKE
Sbjct: 749 IK-SNIGDSAIASYNRMPEWKLFKPFEAAIRDRAYFSQKSKTTPTDEFAISTVAAVLKEK 925
Query: 200 --AWISYQPVLAVMYLVQYMP 218
AW +Y VM ++ ++P
Sbjct: 926 PPAWFTYGHYSTVMAIMYHLP 988
>AW277586
Length = 459
Score = 61.2 bits (147), Expect = 4e-10
Identities = 36/103 (34%), Positives = 52/103 (49%)
Frame = -3
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D I+NA P S +D E L +F VN G + MLR+G G + ++S
Sbjct: 331 IDVGINNAGMSPPMKSFIDTDEADLDLSFAVNAKGVFFGMKHQIRQMLRQGGGIILNVAS 152
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCP 161
AG AP A Y+A+K+A+ G + E KGI+V +CP
Sbjct: 151 VAGLGAAPKLAAYAAAKHAVVGLTKTAAVEYANKGIRVNAICP 23
>TC226411 weakly similar to UP|TRNH_DATST (P50165) Tropinone reductase
homolog (P29X) , partial (83%)
Length = 1109
Score = 58.9 bits (141), Expect = 2e-09
Identities = 46/173 (26%), Positives = 79/173 (45%), Gaps = 2/173 (1%)
Frame = +3
Query: 43 KRKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLT 102
++++++ S+F ++ +++NAA K + D T E + A N L ++
Sbjct: 291 RKRLMEIVGSIFHGK-LNILVNNAATNITKK-ITDYTAEDISAIMGTNFESVYHLCQVAH 464
Query: 103 PFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPG 162
P + G G V +SS AG P +VY+ASK A+N + +L E + I+ V PG
Sbjct: 465 PLLKDSGNGSIVFISSVAGLKALPVFSVYAASKGAMNQFTKNLALEWAKDNIRANAVAPG 644
Query: 163 PIETANNSGSQVPSEKRVSAEKCVELTIIAATHGLKE--AWISYQPVLAVMYL 213
P++T SE S V T + KE A +++ + A Y+
Sbjct: 645 PVKTKLLECIVNSSEGNESINGVVSQTFVGRMGETKEISALVAFLCLPAASYI 803
>TC205954 similar to UP|Q9FK50 (Q9FK50) Brn1-like protein, partial (89%)
Length = 958
Score = 57.8 bits (138), Expect = 5e-09
Identities = 42/126 (33%), Positives = 60/126 (47%), Gaps = 3/126 (2%)
Frame = +1
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAY-ERPKSSVLDVTEESLKATFDVNVFGTITLTRLLT 102
+ + D AE F DS + ++++A + SV D T ES TF VN G R
Sbjct: 298 KSLFDSAERAF-DSPIHILVNSAGVIDGTYPSVADTTVESFDRTFAVNARGAFACAREAA 474
Query: 103 PFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVC-- 160
+ R G G ++++++ PG Y+ASK A+ L EL KG Q+T C
Sbjct: 475 NRLKRGGGGRIILLTTSQVVALRPGYGAYAASKAAVEAMVKILAKEL--KGTQITANCVA 648
Query: 161 PGPIET 166
PGPI T
Sbjct: 649 PGPIAT 666
>BQ453416
Length = 426
Score = 57.8 bits (138), Expect = 5e-09
Identities = 38/130 (29%), Positives = 59/130 (45%), Gaps = 3/130 (2%)
Frame = +2
Query: 40 VTAKRKVVDEAESLFPDSG-VDYMIHNAAYERPK--SSVLDVTEESLKATFDVNVFGTIT 96
VT + +V D G +D M +NA P S++D+ + +N+ G I
Sbjct: 8 VTVEAQVADAVNVAVAHYGKLDIMYNNAGIPGPSIPPSIVDLDLDEFDRVMRINIRGMIA 187
Query: 97 LTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQV 156
+ M+ G G + SS +G G Y+ SK+ + G SL SELC+ GI++
Sbjct: 188 GIKHAARVMIPVGSGSILCTSSISGVLGGLGPHPYTISKFTIPGVVKSLASELCKVGIRI 367
Query: 157 TVVCPGPIET 166
+ P PI T
Sbjct: 368 NCISPAPIPT 397
>TC204815 similar to UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein PM34, partial
(98%)
Length = 1028
Score = 57.4 bits (137), Expect = 6e-09
Identities = 41/147 (27%), Positives = 72/147 (48%), Gaps = 12/147 (8%)
Frame = +3
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
+KVVDE + + +D +++NAA + S+ D+ + L+ F N+F +T+
Sbjct: 447 KKVVDEVINAY--GRIDILVNNAAEQYESDSLEDIDDARLERVFRTNIFSHFFMTKHALK 620
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAV-YSASKYALNGYFHSLRSELCQKGIQVTVVCPG 162
M +G ++ +++ G V Y+++K A+ G+ +L +L KGI+V V PG
Sbjct: 621 HMK---EGSSIINTTSVNAYQGDGTLVDYTSTKGAIVGFTRALALQLVSKGIRVNGVAPG 791
Query: 163 PI-----------ETANNSGSQVPSEK 178
PI ET GS VP ++
Sbjct: 792 PIWTPLIVATMNEETIVRFGSDVPMKR 872
>TC209423 weakly similar to UP|Q9LUE4 (Q9LUE4) Similarity to oxidoreductase,
partial (31%)
Length = 757
Score = 54.7 bits (130), Expect = 4e-08
Identities = 31/99 (31%), Positives = 56/99 (56%)
Frame = +1
Query: 88 DVNVFGTITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRS 147
D+N +G+ T P LR+ KG + ++S G P P ++Y+ASK A+ + +LR
Sbjct: 76 DINFWGSAYGTYFAIPH-LRKSKGKIIAIASCTGWLPVPRMSIYNASKAAVISLYETLRI 252
Query: 148 ELCQKGIQVTVVCPGPIETANNSGSQVPSEKRVSAEKCV 186
EL + I +T+V PG IE+ + G + E ++ +++ +
Sbjct: 253 EL-GRDIGITIVTPGLIESEMSQGKVLFKEGKMVSDQLI 366
>TC221342 similar to UP|Q9SY73 (Q9SY73) F14N23.19, partial (74%)
Length = 792
Score = 54.3 bits (129), Expect = 5e-08
Identities = 40/130 (30%), Positives = 63/130 (47%), Gaps = 3/130 (2%)
Frame = +1
Query: 40 VTAKRKVVDEAESLFPDSGV-DYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLT 98
V++ V + A + + V D +++NA + + +V E A D NV GT +
Sbjct: 301 VSSNENVQEMARVVMDNRSVPDIIVNNAGTINKNNKIWEVPPEDFDAVMDTNVKGTANVL 480
Query: 99 RLLTPFMLRRGKGHFVV--MSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQV 156
R P M+ K V+ MSS G++ A + Y ASK+A+ G S+ E+ +GI V
Sbjct: 481 RHFIPLMIAAKKMEAVIVNMSSGWGRSGAALVSPYCASKWAIEGLSKSVAKEV-PEGIAV 657
Query: 157 TVVCPGPIET 166
+ PG I T
Sbjct: 658 VALNPGVINT 687
>TC217387 similar to GB|AAO24564.1|27808568|BT003132 At3g06060 {Arabidopsis
thaliana;} , partial (92%)
Length = 1545
Score = 53.9 bits (128), Expect = 7e-08
Identities = 34/95 (35%), Positives = 49/95 (50%), Gaps = 4/95 (4%)
Frame = +2
Query: 76 LDVTEES-LKATFDVNVFGTITLTRLLTPFMLRRG---KGHFVVMSSAAGKTPAPGQAVY 131
LD E S +K T DVN+ GT+ L + P M R ++SS AG+ G Y
Sbjct: 557 LDKMELSEVKFTMDVNLMGTLNLIKAALPAMKNRNDPLPASIALVSSQAGQVGIYGYVAY 736
Query: 132 SASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
SASK+ L G SL+ E+ + I V+++ P +T
Sbjct: 737 SASKFGLRGLAESLQQEVIEDNIHVSMIFPPDTDT 841
>TC220612 weakly similar to UP|Q9SBM0 (Q9SBM0) Wts2L, partial (66%)
Length = 1130
Score = 53.9 bits (128), Expect = 7e-08
Identities = 33/94 (35%), Positives = 46/94 (48%)
Frame = +3
Query: 73 SSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYS 132
SS++ + LK F+VNVFG + M+ R G V SSA T Y+
Sbjct: 435 SSIIALDPADLKRVFEVNVFGAFYAAKHAAEIMIPRKIGSIVFTSSAVSVTHPGSPHPYT 614
Query: 133 ASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
ASKYA+ G +L EL + GI+V + P + T
Sbjct: 615 ASKYAVVGLMKNLCVELGKHGIRVNCISPYAVAT 716
>TC206284 similar to UP|Q8S305 (Q8S305) Short-chain alcohol dehydrogenase
(Fragment), partial (94%)
Length = 1100
Score = 52.4 bits (124), Expect = 2e-07
Identities = 41/146 (28%), Positives = 69/146 (47%)
Frame = +2
Query: 38 SKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITL 97
S ++ ++D+ + +D ++ NAA ++L + L +++NV TI L
Sbjct: 272 SSAQQRKNLIDKTVQKY--GKIDVVVSNAAANPSVDAILQTKDSVLDKLWEINVKATILL 445
Query: 98 TRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVT 157
+ P L++G V++SS AG P P A+Y +K AL G +L +E+ +V
Sbjct: 446 LKDAVPH-LQKGSS-VVIISSIAGFNPPPSLAMYGVTKTALLGLTKALAAEMA-PNTRVN 616
Query: 158 VVCPGPIETANNSGSQVPSEKRVSAE 183
V PG + T N S + S V E
Sbjct: 617 CVAPGFVPT--NFASFITSNDAVKKE 688
>TC204814 UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein PM34, complete
Length = 1240
Score = 52.0 bits (123), Expect = 3e-07
Identities = 39/147 (26%), Positives = 69/147 (46%), Gaps = 12/147 (8%)
Frame = +1
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
++VVDE S + +D +++NAA + +V D+ E L+ F N+F + R
Sbjct: 430 KRVVDEVVSAY--GRIDILVNNAAEQYECGTVEDIDEPRLERVFRTNIFSYFFMARHALK 603
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAV-YSASKYALNGYFHSLRSELCQKGIQVTVVCPG 162
M +G ++ +++ + + Y+++K A+ Y L +L KGI+V V PG
Sbjct: 604 HMK---EGSSIINTTSVNAYKGHAKLLDYTSTKGAIVAYTRGLALQLVSKGIRVNGVAPG 774
Query: 163 PI-----------ETANNSGSQVPSEK 178
PI E G+QVP ++
Sbjct: 775 PIWTPLIPASFKEEETAQFGAQVPMKR 855
>TC227493 similar to UP|FABG_CUPLA (P28643) 3-oxoacyl-[acyl-carrier-protein]
reductase, chloroplast precursor (3-ketoacyl-acyl
carrier protein reductase) , partial (83%)
Length = 1452
Score = 51.6 bits (122), Expect = 3e-07
Identities = 41/159 (25%), Positives = 68/159 (41%)
Frame = +1
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
VD +I+NA R ++ + + + D+N+ G T+ M+++ KG V ++S
Sbjct: 643 VDVLINNAGITRD-GLLMRMKKSQWQDVIDLNLTGVFLCTQAAAKIMMKKRKGRIVNIAS 819
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETANNSGSQVPSEK 178
G GQA YSA+K + G ++ E + I V V PG I S + ++
Sbjct: 820 VVGLVGNVGQANYSAAKAGVIGLTKTVAKEYASRNITVNAVAPGFI------ASDMTAKL 981
Query: 179 RVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYM 217
EK + TI +G E LA+ Y+
Sbjct: 982 GQDIEKKILETIPLGRYGQPEEVAGLVEFLALNQAASYI 1098
>TC204757 similar to UP|Q9ASX2 (Q9ASX2) At1g07440/F22G5_16, partial (90%)
Length = 1257
Score = 50.1 bits (118), Expect = 1e-06
Identities = 31/108 (28%), Positives = 52/108 (47%)
Frame = +1
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
++ +++N PK + LDVTEE + N+ L++L P + + + +SS
Sbjct: 340 LNILVNNVGTNVPKHT-LDVTEEDFSFLINTNLESAYHLSQLAHPLLKASEAANIIFISS 516
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
AG + Y A+K A+N +L E + I+ V PGPI+T
Sbjct: 517 IAGVLSIGIGSTYGATKGAMNQLTKNLACEWAKDNIRTNCVAPGPIKT 660
>TC220581 weakly similar to UP|Q9LUE4 (Q9LUE4) Similarity to oxidoreductase,
partial (22%)
Length = 862
Score = 47.8 bits (112), Expect = 5e-06
Identities = 28/79 (35%), Positives = 45/79 (56%)
Frame = +2
Query: 88 DVNVFGTITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRS 147
D+N +G + T P L+ KG +V++S G P P ++Y+ASK A+ +F +LR
Sbjct: 131 DINFWGAVYGTLYAIPH-LKINKGRIIVIASGCGWFPLPRISIYNASKAAVINFFETLRM 307
Query: 148 ELCQKGIQVTVVCPGPIET 166
EL I +T+ PG ++T
Sbjct: 308 EL-GWDIGITIATPGFVKT 361
>TC213428 similar to UP|O42866 (O42866) SPAC3G9.02 protein, partial (7%)
Length = 764
Score = 47.8 bits (112), Expect = 5e-06
Identities = 33/109 (30%), Positives = 49/109 (44%), Gaps = 1/109 (0%)
Frame = +3
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFML-RRGKGHFVVMS 117
++ + NA P SS+LD T VN+ G + + M+ R +G + +
Sbjct: 198 LEILFSNAGIAGPLSSILDFDLNEFDNTMAVNLRGAMAAIKHAARVMVARETRGSIICTT 377
Query: 118 SAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
S AG Y+ASK+ L G S SEL KGI+V + P + T
Sbjct: 378 SVAGSFAGCAGHDYTASKHGLIGLVRSACSELGAKGIRVNSISPYAVAT 524
>TC203383 similar to UP|Q9FXD7 (Q9FXD7) F12A21.14, partial (21%)
Length = 1365
Score = 47.4 bits (111), Expect = 6e-06
Identities = 47/154 (30%), Positives = 68/154 (43%), Gaps = 6/154 (3%)
Frame = +2
Query: 59 VDYMIHNAAYERPKSSVL-DVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFV-VM 116
V +++N P + +V E L VNV GT +T+ + P MLRR KG V +
Sbjct: 557 VGVLVNNVGVSYPYARFFHEVDEGLLNNLIKVNVVGTTKVTQAVLPGMLRRKKGAIVNIG 736
Query: 117 SSAAGKTPA-PGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETANNSGSQVP 175
S AA P+ P AVY+A+K ++ + L E + GI V C P+ A S
Sbjct: 737 SGAAIVIPSDPLYAVYAATKAYIDQFSRCLYVEYKKSGIDVQ--CQIPLYVATKMASIRK 910
Query: 176 SEKRVSAEKCVELTIIAATHGLKEA---WISYQP 206
S + +T G +A WI Y+P
Sbjct: 911 SS-----------FFVPSTDGYAKAGVKWIGYEP 979
>BM519726 similar to GP|22022575|gb| AT3g03330/T21P5_25 {Arabidopsis
thaliana}, partial (7%)
Length = 437
Score = 47.0 bits (110), Expect = 8e-06
Identities = 30/79 (37%), Positives = 41/79 (50%), Gaps = 4/79 (5%)
Frame = +2
Query: 93 GTITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKY----ALNGYFHSLRSE 148
GTITLT+LL PFML+RG GHFVV+ + G ++ A Y + YFH R
Sbjct: 113 GTITLTKLLAPFMLKRGHGHFVVVCRNSLIISYSGYGIWMAIVY*VCFCFHYYFHVERIS 292
Query: 149 LCQKGIQVTVVCPGPIETA 167
+ + C G +E+A
Sbjct: 293 M-----DFFLSCLGALESA 334
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,658,005
Number of Sequences: 63676
Number of extensions: 150114
Number of successful extensions: 812
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 807
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 809
length of query: 254
length of database: 12,639,632
effective HSP length: 95
effective length of query: 159
effective length of database: 6,590,412
effective search space: 1047875508
effective search space used: 1047875508
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147179.10


