

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.8 + phase: 0
(461 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
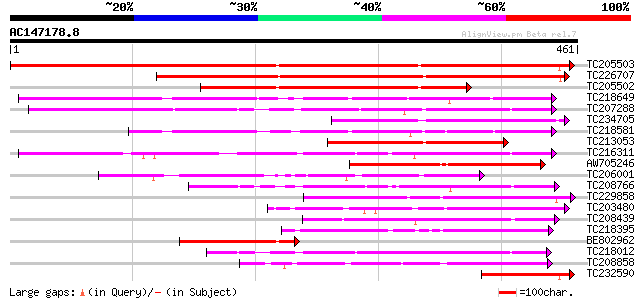
Score E
Sequences producing significant alignments: (bits) Value
TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete 439 e-123
TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 304 6e-83
TC205502 homologue to UP|Q7XA50 (Q7XA50) Sorbitol-like transport... 205 3e-53
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 190 1e-48
TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete 152 4e-37
TC234705 weakly similar to GB|CAD70577.1|32698459|MMU549317 solu... 130 2e-30
TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F2... 127 8e-30
TC213053 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 127 1e-29
TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete 125 4e-29
AW705246 111 8e-25
TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, parti... 108 4e-24
TC208766 similar to UP|STA_RICCO (Q10710) Sugar carrier protein ... 99 3e-21
TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporte... 99 5e-21
TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, par... 94 1e-19
TC208439 weakly similar to UP|O04078 (O04078) Monosaccharid tran... 94 1e-19
TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like pr... 92 5e-19
BE802962 similar to GP|29691859|gb| sorbitol transporter {Malus ... 89 4e-18
TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporte... 88 7e-18
TC208858 similar to UP|Q9M9Z8 (Q9M9Z8) F20B17.24, partial (51%) 77 2e-14
TC232590 homologue to UP|Q7XA50 (Q7XA50) Sorbitol-like transport... 75 6e-14
>TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete
Length = 1928
Score = 439 bits (1128), Expect = e-123
Identities = 224/472 (47%), Positives = 325/472 (68%), Gaps = 13/472 (2%)
Frame = +1
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA ++IK +L++SD Q+++L GI+N +L +AGR SD+IGRRYTI L F +G
Sbjct: 319 MSGAAIYIKRDLKVSDEQIEILLGIINLYSLIGSCLAGRTSDWIGRRYTIGLGGAIFLVG 498
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
S LMG+ + LM GR +AG G+G+AL+I VY+AE+S S RGFLTS P++ IN G L
Sbjct: 499 STLMGFYPHYSFLMCGRFVAGIGIGYALMIAPVYTAEVSPASSRGFLTSFPEVFINGGIL 678
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNY KL+L++GWR+ML + +IPS+ L +L + ESPRWLVM+GRLG+A+KVL
Sbjct: 679 LGYISNYGFSKLTLKVGWRMMLGVGAIPSVVLTEGVLAMPESPRWLVMRGRLGEARKVLN 858
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+SK+EA+ R+ EIK A GI E+C ++V V+K++ +G G KE+F P+P + I+I
Sbjct: 859 KTSDSKEEAQLRLAEIKQAAGIPESCNDDVVQVNKQS-NGEGVWKELFLYPTPAIRHIVI 1035
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AA+G+H FQ GV+ + LYSPRIF + GIT+ LLATV +G +T+F L + F LD+
Sbjct: 1036AALGIHFFQQASGVDAVVLYSPRIFEKAGITNDTHKLLATVAVGFVKTVFILAATFTLDR 1215
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LLL S GG++ S+L L + ++++S+ + +WA+ +I ++ +IG G
Sbjct: 1216VGRRPLLLSSVGGMVLSLLTLAISLTVIDHSE--RKLMWAVGSSIAMVLAYVATFSIGAG 1389
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+EIFPLRLRAQG V +NR T+ V +F+S+ + IT+GG FFL+ G +
Sbjct: 1390PITWVYSSEIFPLRLRAQGAAAGVAVNRTTSAVVSMTFLSLTRAITIGGAFFLYCGIATV 1569
Query: 421 GWWFYYSFLPETKGRSLEDMETIFG------------KNSNSEV-QMKYGSN 459
GW F+Y+ LPET+G++LEDME FG +N N +V Q++ G+N
Sbjct: 1570GWIFFYTVLPETRGKTLEDMEGSFGTFRSKSNASKAVENENGQVAQVQLGTN 1725
>TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 2338
Score = 304 bits (778), Expect = 6e-83
Identities = 154/347 (44%), Positives = 228/347 (65%), Gaps = 11/347 (3%)
Frame = +3
Query: 120 LLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVL 179
L+GY+SNY KL+LRLGWR+ML + +IPSI + + +L + ESPRWLV +GRLG+AK+VL
Sbjct: 6 LIGYISNYGFSKLALRLGWRLMLGVGAIPSILIGVAVLAMPESPRWLVAKGRLGEAKRVL 185
Query: 180 LLISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRIL 239
IS S++EA R+ +IK+ GI ++C ++V VSK+T G G +E+F P+P V I
Sbjct: 186 YKISESEEEARLRLADIKDTAGIPQDCDDDVVLVSKQTH-GHGVWRELFLHPTPAVRHIF 362
Query: 240 IAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLD 299
IA++G+H F G++ + LYSPRIF + GI LLATV +G +T+ L++ F LD
Sbjct: 363 IASLGIHFFAQATGIDAVVLYSPRIFEKAGIKSDNYRLLATVAVGFVKTVSILVATFFLD 542
Query: 300 KIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGI 359
+ GRR+LLL S G+I S+L L + +V++S+ WA+ +I + +IG
Sbjct: 543 RAGRRVLLLCSVSGLILSLLTLGLSLTVVDHSQTTLN--WAVGLSIAAVLSYVATFSIGS 716
Query: 360 GAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNV 419
G +TWVYS+EIFPLRLRAQG+ + +NR+T+ + +F+S+ K IT+GG FFLF G
Sbjct: 717 GPITWVYSSEIFPLRLRAQGVAIGAAVNRVTSGVIAMTFLSLQKAITIGGAFFLFAGVAA 896
Query: 420 LGWWFYYSFLPETKGRSLEDMETIFGK-----------NSNSEVQMK 455
+ W F+Y+ LPET+G++LE++E FG + N E+Q+K
Sbjct: 897 VAWIFHYTLLPETRGKTLEEIEKSFGNFCRKPKAEEGLDDNVEIQLK 1037
>TC205502 homologue to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(42%)
Length = 659
Score = 205 bits (522), Expect = 3e-53
Identities = 104/220 (47%), Positives = 152/220 (68%)
Frame = +1
Query: 156 MLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSK 215
+L + ESPRWLVM+GRLG+A+KVL S+S++EA+ R+ EIK A GI E+C ++V V+K
Sbjct: 7 VLAMPESPRWLVMRGRLGEARKVLNKTSDSREEAQLRLAEIKQAAGIPESCNDDVVQVTK 186
Query: 216 KTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGT 275
++ +G G KE+F P+P + I+IAA+G+H FQ GV+ + LYSPRIF + GI D
Sbjct: 187 RS-TGEGVWKELFLYPTPPIRHIVIAALGIHFFQQASGVDAVVLYSPRIFEKAGIKDDTH 363
Query: 276 LLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGE 335
LLATV +G +T+F L + F LD++GRR LLL S GG++ S+L L + I+ +S+
Sbjct: 364 KLLATVAVGFVKTVFILAATFTLDRVGRRPLLLSSVGGMVLSLLTLAISLTIIGHSE--R 537
Query: 336 EPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRL 375
+ +WA+ +I ++ +IG G +TWVYS+EIFPLRL
Sbjct: 538 KLMWAVALSIAMVLAYVATFSIGAGPITWVYSSEIFPLRL 657
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 190 bits (482), Expect = 1e-48
Identities = 131/439 (29%), Positives = 224/439 (50%), Gaps = 2/439 (0%)
Frame = +2
Query: 8 IKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYG 67
I +L +S + L + N A+ + +G++++YIGR+ ++M++SI +G + + +
Sbjct: 257 IINDLGLSVSEFSLFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIASIPNIIGWLAISFA 436
Query: 68 SSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNY 127
L +GR + GFGVG V VY AEIS P+ RG L S+ LS+ IG +L YL
Sbjct: 437 KDSSFLYMGRLLEGFGVGIISYTVPVYIAEISPPNLRGGLVSVNQLSVTIGIMLAYL--- 607
Query: 128 FLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQ 187
L + + WRI+ I +P L+ + + ESPRWL G + + L ++
Sbjct: 608 ----LGIFVEWRILAIIGILPCTILIPALFFIPESPRWLAKMGMTEEFETSLQVLRGFDT 775
Query: 188 EAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHI 247
+ + EIK AV N + K R ++ P L+ IG+ I
Sbjct: 776 DISVEVNEIKRAVA-STNTRITVRFADLKQR--------RYWLP-------LMIGIGLLI 907
Query: 248 FQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILL 307
Q + G+ G+ YS IF GI+ AT G+G Q L T L+ +L DK GRR+LL
Sbjct: 908 LQQLSGINGVLFYSSTIFRNAGISSSDA---ATFGVGAVQVLATSLTLWLADKSGRRLLL 1078
Query: 308 LVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNA--IGIGAVTWV 365
+VS+ G+ FS+L + + + ++ S L+ I+ T+ ++ ++A A +G+GA+ W+
Sbjct: 1079IVSATGMSFSLLVVAI-TFYIKASISETSSLYGILSTLSLVGVVAMVIAFSLGMGAMPWI 1255
Query: 366 YSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFY 425
+EI P+ ++ V + N + + +VT ++ + GGTF ++ L F
Sbjct: 1256IMSEILPINIKGLAGSVATLANWLFS-WLVTLTANMLLDWSSGGTFTIYAVVCALTVVFV 1432
Query: 426 YSFLPETKGRSLEDMETIF 444
++PETKG+++E+++ F
Sbjct: 1433TIWVPETKGKTIEEIQWSF 1489
>TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete
Length = 1813
Score = 152 bits (383), Expect = 4e-37
Identities = 116/431 (26%), Positives = 201/431 (45%), Gaps = 2/431 (0%)
Frame = +1
Query: 16 DMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMI 75
D + L L AL A L+ GR+ +I++ ++ F G+IL + +L+I
Sbjct: 271 DQVLTLFTSSLYFSALVMTFFASFLTRKKGRKASIIVGALSFLAGAILNAAAKNIAMLII 450
Query: 76 GRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLR 135
GR + G G+GF V +Y +E++ RG + L + G L+ L NYF K+
Sbjct: 451 GRVLLGGGIGFGNQAVPLYLSEMAPAKNRGAVNQLFQFTTCAGILIANLVNYFTEKIH-P 627
Query: 136 LGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKE 195
GWRI L + +P+ +++ + E+P LV QGRL AK+VL I + + E ++
Sbjct: 628 YGWRISLGLAGLPAFAMLVGGICCAETPNSLVEQGRLDKAKQVLQRIRGT-ENVEAEFED 804
Query: 196 IKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVE 255
+K A ++ +S L + Y+P +++I A+G+ FQ + G
Sbjct: 805 LKEA-----------SEEAQAVKSPFRTLLKRKYRP-----QLIIGALGIPAFQQLTGNN 936
Query: 256 GIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVI 315
I Y+P IF +G +L + + G + + T++S FL+DK GRR L + +I
Sbjct: 937 SILFYAPVIFQSLGFGANASLFSSFITNG-ALLVATVISMFLVDKYGRRKFFLEAGFEMI 1113
Query: 316 FSML--GLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPL 373
M+ G + ++G+ F ++VI++ G + W+ +E+FPL
Sbjct: 1114CCMIITGAVLAVNFGHGKEIGK---GVSAFLVVVIFLFVLAYGRSWGPLGWLVPSELFPL 1284
Query: 374 RLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETK 433
+R+ + V +N I V F+ + G F LF + +F + LPETK
Sbjct: 1285EIRSSAQSIVVCVNMIFTALVAQLFLMSLCHLKF-GIFLLFASLIIFMSFFVFFLLPETK 1461
Query: 434 GRSLEDMETIF 444
+E++ +F
Sbjct: 1462KVPIEEIYLLF 1494
>TC234705 weakly similar to GB|CAD70577.1|32698459|MMU549317 solute carrier
family 2 (facilitated glucose transporter), member 12
{Mus musculus;} , partial (5%)
Length = 992
Score = 130 bits (326), Expect = 2e-30
Identities = 70/194 (36%), Positives = 115/194 (59%)
Frame = +1
Query: 262 PRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGL 321
P IF GI D LL ATV +G+++T+F L++ L+DK+GR+ LL++S+ G+ + +
Sbjct: 1 PEIFQAAGIEDNSKLLAATVAVGVAKTIFILVAIILIDKLGRKPLLMISTIGMTVCLFCM 180
Query: 322 CVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLG 381
A++ +AI I+ + F ++G+G V WV ++EIFPLR+RAQ
Sbjct: 181 GATLALLGKGS------FAIALAILFVCGNVAFFSVGLGPVCWVLTSEIFPLRVRAQASA 342
Query: 382 VCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDME 441
+ + NR+ + V SF+S+ + I++ GTFF+F + L F + +PETKG+SLE +E
Sbjct: 343 LGAVANRVCSGLVAMSFLSVSEAISVAGTFFVFAAISALAIAFVVTLVPETKGKSLEQIE 522
Query: 442 TIFGKNSNSEVQMK 455
+F + E+Q K
Sbjct: 523 MMF--QNEYEIQGK 558
>TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;} , partial (66%)
Length = 1340
Score = 127 bits (320), Expect = 8e-30
Identities = 97/352 (27%), Positives = 169/352 (47%), Gaps = 4/352 (1%)
Frame = +2
Query: 97 EISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILM 156
EI+ G L S+ LSI IG +L YL L L + WRI+ + +P L+ +
Sbjct: 5 EIAPNHLIGGLGSVNQLSITIGIMLAYL-------LGLFVNWRILAILGILPCTVLIPGL 163
Query: 157 LQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKK 216
+ ESPRWL G + + + L ++ + + EIK +V K+
Sbjct: 164 FFIPESPRWLAKMGMIDEFETSLQVLRGFDTDISVEVHEIKRSVA----------STGKR 313
Query: 217 TRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTL 276
LK Y + L+ IG+ + Q + G+ GI YS IF GI+
Sbjct: 314 AAIRFADLKRKRY------WFPLMVGIGLLVLQQLSGINGILFYSTTIFANAGISSSEA- 472
Query: 277 LLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVC----SAIVENSK 332
ATVG+G Q + T +S +L+DK GRR+LL++SS + S+L + + + E+S
Sbjct: 473 --ATVGLGAVQVIATGISTWLVDKSGRRLLLIISSSVMTVSLLIVSIAFYLEGVVSEDSH 646
Query: 333 LGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNV 392
L + I+ + ++ ++ GF ++G+G + W+ +EI P+ ++ + + N + +
Sbjct: 647 LFS--ILGIVSIVGLVAMVIGF-SLGLGPIPWLIMSEILPVNIKGLAGSIATMGNWLISW 817
Query: 393 AVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDMETIF 444
+T ++ + GGTF ++ F ++PETKGR+LE+++ F
Sbjct: 818 G-ITMTANLLLNWSSGGTFTIYTVVAAFTIAFIAMWVPETKGRTLEEIQFSF 970
>TC213053 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(27%)
Length = 440
Score = 127 bits (318), Expect = 1e-29
Identities = 64/147 (43%), Positives = 100/147 (67%)
Frame = +1
Query: 259 LYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSM 318
LYSP IF + G+ G LLATV +G ++T+F L++ FLLD++GRR LLL S GG++FS+
Sbjct: 4 LYSPEIFKKAGLESDGEQLLATVAVGFAKTVFILVATFLLDRVGRRPLLLTSVGGMVFSL 183
Query: 319 LGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQ 378
L L + ++++S+ + WAI +I ++ ++G G +TWVYS+EIFPLRLRAQ
Sbjct: 184 LTLGLSLTVIDHSRAVLK--WAIGLSIGMVLSYVSTFSVGAGPITWVYSSEIFPLRLRAQ 357
Query: 379 GLGVCVIMNRITNVAVVTSFISIYKTI 405
G + V++NR+T+ + +F+S+ I
Sbjct: 358 GAAMGVVVNRVTSGVISMTFLSLSNKI 438
>TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete
Length = 1925
Score = 125 bits (314), Expect = 4e-29
Identities = 104/450 (23%), Positives = 201/450 (44%), Gaps = 13/450 (2%)
Frame = +1
Query: 8 IKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYG 67
+ + + + + L AL + ++A ++ GR+ +++ + F +G+++ G+
Sbjct: 307 VNQYCQYDSQTLTMFTSSLYLAALLSSLVASTVTRRFGRKLSMLFGGLLFLVGALINGFA 486
Query: 68 SSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFL---TSLPDLSIN----IGFL 120
+L++GR + GFG+GFA + + + P GF+ S+ L+ N G
Sbjct: 487 QHVWMLIVGRILLGFGIGFANQVCA------TLPI*NGFIQI*RSIGTLAFNCQSQFGIP 648
Query: 121 LGYLSNYFLG-KLSLRLGWRI-MLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKV 178
G L K + GW+I + +P++ + + L L ++P ++ +G
Sbjct: 649 RGQCVELLLWLKSMVAWGWKIEVWEGAMVPALIITVGSLVLPDTPNSMIERG-------- 804
Query: 179 LLLISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRI 238
+++A+ +++ I+ +DE + ++ + K PH
Sbjct: 805 ------DREKAKAQLQRIRGIDNVDEEFNDLVAASESSSQVEHPWRNLLQRKYRPH---- 954
Query: 239 LIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLL 298
L A+ + FQ + G+ I Y+P +F +G D L+ A + G+ + T +S + +
Sbjct: 955 LTMAVLIPFFQQLTGINVIMFYAPVLFSSIGFKDDAALMSAVI-TGVVNVVATCVSIYGV 1131
Query: 299 DKIGRRILLLVSSGGVIFSMLGLCVCSAI----VENSKLGEEPLWAIIFTIIVIYIMAGF 354
DK GRR L L GGV + V +AI + G+ P W I ++ I I
Sbjct: 1132DKWGRRALFL--EGGVQMLICQAVVAAAIGAKFGTDGNPGDLPKWYAIVVVLFICIYVSA 1305
Query: 355 NAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLF 414
A G + W+ +EIFPL +R+ + V +N + + F+++ + G F F
Sbjct: 1306FAWSWGPLGWLVPSEIFPLEIRSAAQSINVSVNMLFTFLIAQVFLTMLCHMKF-GLFLFF 1482
Query: 415 VGTNVLGWWFYYSFLPETKGRSLEDMETIF 444
++ +F Y FLPETKG +E+M ++
Sbjct: 1483AFFVLIMTFFVYFFLPETKGIPIEEMGQVW 1572
>AW705246
Length = 484
Score = 111 bits (277), Expect = 8e-25
Identities = 55/159 (34%), Positives = 97/159 (60%)
Frame = +2
Query: 277 LLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEE 336
+L +V +G +T+ ++ LD+ GRR+LLL + G+I S++ L + +V++S+
Sbjct: 2 VLTSVAVGFDKTVCIWVATIFLDRAGRRVLLLCNVSGLILSLMTLGLSLTVVDHSQTTLN 181
Query: 337 PLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVT 396
+ ++ Y+ A F+ I G +TWVY +EIFPLRL A G+ + +N++T+ +
Sbjct: 182 CAVGLRIDSVLSYV-ATFS-IWSGPITWVYISEIFPLRLWAHGVAIGAAVNKVTSGVIAM 355
Query: 397 SFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGR 435
+F+ + K IT+GG FFLF G + W F+Y+ +PE +G+
Sbjct: 356 TFLYLQKAITIGGAFFLFAGVAAVAWIFHYTLIPEARGK 472
>TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, partial (66%)
Length = 951
Score = 108 bits (271), Expect = 4e-24
Identities = 91/320 (28%), Positives = 145/320 (44%), Gaps = 6/320 (1%)
Frame = +3
Query: 73 LMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSI----NIGFLLGYLSNYF 128
L +GR G+G+G +V VY AEI+ + RG L + L I ++ FLLG + N
Sbjct: 39 LDMGRFFTGYGIGVISYVVPVYIAEIAPKNLRGGLATTNQLLIVTGGSVSFLLGSVIN-- 212
Query: 129 LGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQE 188
WR + +P I L++ + + ESPRWL GR + + L + +
Sbjct: 213 ---------WRELALAGLVPCICLLVGLCFIPESPRWLAKVGREKEFQLALSRLRGKHAD 365
Query: 189 AEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIF 248
EI + + E+ KT+ L ++F S +V+ ++I +G+
Sbjct: 366 ISDEAAEILDYIETLESLP--------KTK-----LLDLFQ--SKYVHSVVIG-VGLMAC 497
Query: 249 QNICGVEGIFLYSPRIFGRMGITD--KGTLLLATVGIGISQTLFTLLSCFLLDKIGRRIL 306
Q G+ GI Y+ IF G++ GT+ A + Q FTLL L+DK GRR L
Sbjct: 498 QQSVGINGIGFYTAEIFVAAGLSSGKAGTIAYACI-----QIPFTLLGAILMDKSGRRPL 662
Query: 307 LLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVY 366
++VS+ G + + S L P W I + I +IG+G+V WV
Sbjct: 663 VMVSAAGTFLGCFVAAFAFFLKDQSLL---PEWVPILAFAGVLIYIAAFSIGLGSVPWVI 833
Query: 367 STEIFPLRLRAQGLGVCVIM 386
+EIFP+ L+ + V++
Sbjct: 834 MSEIFPIHLKGTAGSLVVLV 893
>TC208766 similar to UP|STA_RICCO (Q10710) Sugar carrier protein A, partial
(55%)
Length = 1202
Score = 99.4 bits (246), Expect = 3e-21
Identities = 78/305 (25%), Positives = 151/305 (48%), Gaps = 3/305 (0%)
Frame = +1
Query: 146 SIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAVGIDEN 205
++P++ + + + L ++P L+ +G +K+L I +K E + +++ +A
Sbjct: 1 AVPALLMTVGGIFLPDTPNSLIERGLAEKGRKLLEKIRGTK-EVDAEFQDMVDA----SE 165
Query: 206 CTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIF 265
++I H + + E Y+P L+ AI + FQ + G+ I Y+P +F
Sbjct: 166 LAKSIKHPFRN-------ILERRYRPE------LVMAIFMPTFQILTGINSILFYAPVLF 306
Query: 266 GRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVCS 325
MG +L+ + + G+ + T +S +D++GRR+LL+ SGG+ M+ +
Sbjct: 307 QSMGFGGDASLISSALTGGVLASS-TFISIATVDRLGRRVLLV--SGGL--QMITCQIIV 471
Query: 326 AIVENSKLGEEPLWAIIFTIIVIYIMAGFNAI---GIGAVTWVYSTEIFPLRLRAQGLGV 382
AI+ K G + + F+I+V+ ++ F G + W +EIFPL +R+ G G+
Sbjct: 472 AIILGVKFGADQELSKGFSILVVVVICLFVVAFGWSWGPLGWTVPSEIFPLEIRSAGQGI 651
Query: 383 CVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDMET 442
V +N + + +F+++ + G F F G + F Y FLPETKG +E+M
Sbjct: 652 TVAVNLLFTFIIAQAFLALLCSFKFG-IFLFFAGWITIMTIFVYLFLPETKGIPIEEMSF 828
Query: 443 IFGKN 447
++ ++
Sbjct: 829 MWRRH 843
>TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporter 10 (Mus
musculus 13 days embryo heart cDNA, RIKEN full-length
enriched library, clone:D330001H08 product:solute
carrier family 2 (facilitated glucose transporter),
member 10, full insert sequence), partial (4%)
Length = 715
Score = 98.6 bits (244), Expect = 5e-21
Identities = 67/226 (29%), Positives = 113/226 (49%), Gaps = 5/226 (2%)
Frame = +3
Query: 240 IAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLD 299
+ G+ FQ G+ + YSP I G LL ++ + T+L +L+D
Sbjct: 21 LVGAGLLAFQQFTGINTVMYYSPTIVQMAGFHANELALLLSLIVAGMNAAGTILGIYLID 200
Query: 300 KIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGI 359
GR+ L L S GGVI S++ L A + S E + + +YI GF + G+
Sbjct: 201 HAGRKKLALSSLGGVIVSLVILAF--AFYKQSSTSNELYGWLAVVGLALYI--GFFSPGM 368
Query: 360 GAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNV 419
G V W S+EI+P R G+ + ++N+ V +F+SI + I +G TF + V
Sbjct: 369 GPVPWTLSSEIYPEEYRGICGGMSATVCWVSNLIVSETFLSIAEGIGIGSTFLIIGVIAV 548
Query: 420 LGWWFYYSFLPETKGRSLEDMETI-----FGKNSNSEVQMKYGSNN 460
+ + F ++PETKG + +++E I +GKN N++ ++ GS++
Sbjct: 549 VAFVFVLVYVPETKGLTFDEVEVIWRERAWGKNPNTQNLLEQGSSS 686
>TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (38%)
Length = 1040
Score = 94.4 bits (233), Expect = 1e-19
Identities = 71/259 (27%), Positives = 118/259 (45%), Gaps = 13/259 (5%)
Frame = +2
Query: 210 IVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMG 269
+VH S+ T S G + K + P V L+ +G+ I Q G+ G+ Y+P+I G
Sbjct: 134 MVHPSE-TASKGPSWKALL---EPGVKHALVVGVGIQILQQFSGINGVLYYTPQILEEAG 301
Query: 270 ITDKGTLLLATVGIGISQ--------TLFTLLSCF-----LLDKIGRRILLLVSSGGVIF 316
+ +LL+ +GIG T F +L C L+D GRR LLL + +I
Sbjct: 302 VE----VLLSDIGIGSESASFLISAFTTFLMLPCIGVAMKLMDVSGRRQLLLTTIPVLIV 469
Query: 317 SMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLR 376
S++ L + S + G AI +V+Y +G G + + +EIFP R+R
Sbjct: 470 SLIILVIGSLV----NFGNVAHAAISTVCVVVYFCCF--VMGYGPIPNILCSEIFPTRVR 631
Query: 377 AQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRS 436
+ +C ++ I ++ + S + ++ LGG F ++ + W F + +PETKG
Sbjct: 632 GLCIAICALVFWIGDIIITYSLPVMLGSLGLGGVFAIYAVVCFISWIFVFLKVPETKGMP 811
Query: 437 LEDMETIFGKNSNSEVQMK 455
LE + F + K
Sbjct: 812 LEVISEFFSVGAKQAASAK 868
>TC208439 weakly similar to UP|O04078 (O04078) Monosaccharid transport
protein, partial (43%)
Length = 936
Score = 94.0 bits (232), Expect = 1e-19
Identities = 65/212 (30%), Positives = 102/212 (47%), Gaps = 3/212 (1%)
Frame = +1
Query: 239 LIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLL 298
L+ AI + FQ G+ I Y+P +F +G G L++ V IG + + TL+S L+
Sbjct: 10 LVFAICIPFFQQFTGLNVITFYAPILFRTIGF-GSGASLMSAVIIGSFKPVSTLVSILLV 186
Query: 299 DKIGRRILLLVSSGGVIFSMLGLCVCSAIV--ENSKLGEEPLWAIIFTIIVIYIMAGFNA 356
DK GRR L L ++ + + + A+ N G P W I + +I + A
Sbjct: 187 DKFGRRTLFLEGGAQMLICQIIMTIAIAVTFGTNGNPGTLPKWYAIVVVGIICVYVSGFA 366
Query: 357 IGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVG 416
G + W+ +EIFPL +R + V +N I+ + F S+ + G F+F G
Sbjct: 367 WSWGPLGWLIPSEIFPLEIRPAAQSITVGVNMISTFFIAQFFTSMLCHMKFG--LFIFFG 540
Query: 417 T-NVLGWWFYYSFLPETKGRSLEDMETIFGKN 447
V+ F Y LPETKG LE+M ++ K+
Sbjct: 541 CFVVIMTTFIYKLLPETKGIPLEEMSMVWQKH 636
>TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like protein,
partial (40%)
Length = 1013
Score = 92.0 bits (227), Expect = 5e-19
Identities = 68/221 (30%), Positives = 108/221 (48%)
Frame = +1
Query: 222 GALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATV 281
G E+F P+ + I G+ +FQ I G + Y+ I G + +V
Sbjct: 82 GNFLEVFQGPN---LKAFIIGGGLVLFQQITGQPSVLYYAGPILQSAGFSAASDATKVSV 252
Query: 282 GIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAI 341
IG+ + L T ++ +D +GRR LL+ GGV L L + SA + LG PL A+
Sbjct: 253 VIGLFKLLMTWIAVLKVDDLGRRPLLI---GGVSGIALSLVLLSAYYKF--LGGFPLVAV 417
Query: 342 IFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISI 401
+++Y+ G I G ++W+ +E+FPLR R +G+ + V+ N +N V +F +
Sbjct: 418 --GALLLYV--GCYQISFGPISWLMVSEVFPLRTRGKGISLAVLTNFASNAVVTFAFSPL 585
Query: 402 YKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDMET 442
+ + F LF L F +PETKG SLED+E+
Sbjct: 586 KEFLGAENLFLLFGAIATLSLLFIIFSVPETKGMSLEDIES 708
>BE802962 similar to GP|29691859|gb| sorbitol transporter {Malus x
domestica}, partial (19%)
Length = 295
Score = 89.0 bits (219), Expect = 4e-18
Identities = 46/97 (47%), Positives = 66/97 (67%)
Frame = +2
Query: 139 RIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKN 198
R+ML + +IPS+ L + +L + ESPRWLVM+GRLG+A+ VL S SK+EA+ R+ EIK
Sbjct: 2 RMMLGVGAIPSVLLTVGVLAMPESPRWLVMRGRLGEARNVLNKSSYSKEEAQLRLAEIKQ 181
Query: 199 AVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHV 235
A GI E+C ++V V + G G KE+F P+P +
Sbjct: 182 AAGIPESCNDDVVQVYNHS-YGEGVWKELFLYPTPAI 289
>TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporter 4, partial
(54%)
Length = 1142
Score = 88.2 bits (217), Expect = 7e-18
Identities = 70/280 (25%), Positives = 123/280 (43%)
Frame = +3
Query: 161 ESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSG 220
++P L+ +GRL + K VL I + E +E+ A V+K+ +
Sbjct: 3 DTPNSLIERGRLEEGKTVLKKIRGT-DNIELEFQELVEAS-----------RVAKEVKHP 146
Query: 221 GGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLAT 280
L + +P L+ +I + IFQ G+ I Y+P +F +G + +L A
Sbjct: 147 FRNLLKRRNRPQ------LVISIALQIFQQFTGINAIMFYAPVLFNTLGFKNDASLYSAV 308
Query: 281 VGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWA 340
+ G L T++S + +DK+GRR+LLL + + S + + + I +
Sbjct: 309 I-TGAVNVLSTVVSIYSVDKLGRRMLLLEAGVQMFLSQVVIAIILGIKVTDHSDDLSKGI 485
Query: 341 IIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFIS 400
I ++++ A G + W+ +E FPL R+ G V V +N + + +F+S
Sbjct: 486 AILVVVMVCTFVSSFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNLLFTFVIAQAFLS 665
Query: 401 IYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDM 440
+ G F F G ++ F LPETK +E+M
Sbjct: 666 MLCHFKF-GIFLFFSGWVLVMSVFVLFLLPETKNVPIEEM 782
>TC208858 similar to UP|Q9M9Z8 (Q9M9Z8) F20B17.24, partial (51%)
Length = 993
Score = 76.6 bits (187), Expect = 2e-14
Identities = 65/257 (25%), Positives = 119/257 (46%), Gaps = 3/257 (1%)
Frame = +3
Query: 188 EAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGG---ALKEMFYKPSPHVYRILIAAIG 244
EAE +++ V + T+ +SK R G L E+ Y +R++
Sbjct: 9 EAEAAFEKLLGGVHVKPAMTE----LSKSDRGDGSDSVKLSELIYG---RYFRVMFIGST 167
Query: 245 VHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRR 304
+ Q + G+ +F +S +F G+ +A +G+ L ++++ L+DK+GR+
Sbjct: 168 LFALQQLSGINAVFYFSSTVFESFGVPSD----IANSCVGVCNLLGSVVAMILMDKLGRK 335
Query: 305 ILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTW 364
+LLL S G+ SM GL V +A S G L + V+ A G G V
Sbjct: 336 VLLLGSFLGMGLSM-GLQVIAASSFASGFGSMYLSVGGMLLFVLSF-----AFGAGPVPC 497
Query: 365 VYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWF 424
+ +EI P +RA+ + +C+ ++ + N V F+ + + I + +F ++ F
Sbjct: 498 LIMSEILPSNIRAKAMAICLAVHWVINFFVGLFFLRLLELIGAQLLYSIFGFCCLIAVVF 677
Query: 425 YYSFLPETKGRSLEDME 441
+ ETKG+SL+++E
Sbjct: 678 VKKNILETKGKSLQEIE 728
>TC232590 homologue to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(18%)
Length = 403
Score = 75.1 bits (183), Expect = 6e-14
Identities = 38/89 (42%), Positives = 57/89 (63%), Gaps = 13/89 (14%)
Frame = +2
Query: 384 VIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDMETI 443
V++NR T+ V +F+S+ + IT+GG FFL+ G LGW F+Y+ LPET+G++LEDME
Sbjct: 23 VVVNRTTSGVVSMTFLSLSEAITIGGAFFLYCGIATLGWIFFYTLLPETRGKTLEDMEGS 202
Query: 444 FG------------KNSNSEV-QMKYGSN 459
FG +N N +V Q++ G+N
Sbjct: 203 FGTFRSKSNATKGVENGNGQVAQVQLGTN 289
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.144 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,937,241
Number of Sequences: 63676
Number of extensions: 313383
Number of successful extensions: 2090
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 2020
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2043
length of query: 461
length of database: 12,639,632
effective HSP length: 101
effective length of query: 360
effective length of database: 6,208,356
effective search space: 2235008160
effective search space used: 2235008160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147178.8


