

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147013.5 - phase: 0
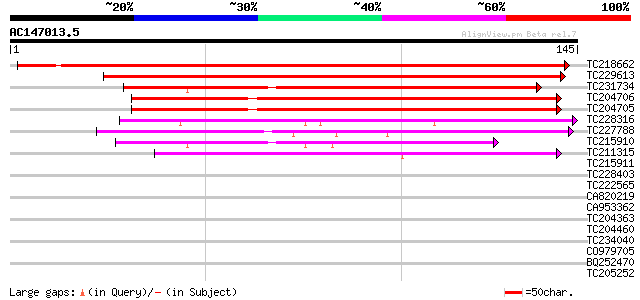
(145 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218662 similar to UP|Q9FKW8 (Q9FKW8) Senescence-associated pro... 204 2e-53
TC229613 weakly similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (40%) 176 5e-45
TC231734 weakly similar to UP|Q7Y234 (Q7Y234) At2g21045, partial... 120 3e-28
TC204706 similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (61%) 108 7e-25
TC204705 similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (61%) 108 1e-24
TC228316 similar to UP|Q94A65 (Q94A65) AT4g27700/T29A15_190, par... 47 4e-06
TC227788 weakly similar to UP|Q8LBR6 (Q8LBR6) Rhodanese-like fam... 44 4e-05
TC215910 similar to UP|O48529 (O48529) Rhodanese-like family pro... 42 8e-05
TC211315 similar to UP|Q9LMU3 (Q9LMU3) F2H15.8 protein, partial ... 40 5e-04
TC215911 similar to UP|Q6H444 (Q6H444) Rhodanese family protein-... 37 0.003
TC228403 similar to UP|Q6ZI49 (Q6ZI49) Rhodanese-like domain-con... 30 0.53
TC222565 similar to UP|Q7XJN9 (Q7XJN9) At2g40760 protein, partia... 29 0.69
CA820219 weakly similar to GP|20197120|gb| expressed protein {Ar... 28 1.2
CA953362 28 2.0
TC204363 similar to UP|O22403 (O22403) ADL2 protein, partial (15%) 28 2.0
TC204460 homologue to UP|B2_DAUCA (P37707) B2 protein, partial (... 27 3.4
TC234040 27 3.4
CO979705 27 4.5
BQ252470 27 4.5
TC205252 homologue to UP|Q9ZP50 (Q9ZP50) FtsH-like protein Pftf ... 26 5.9
>TC218662 similar to UP|Q9FKW8 (Q9FKW8) Senescence-associated protein
sen1-like protein (AT5g66170/K2A18_25), partial (18%)
Length = 941
Score = 204 bits (518), Expect = 2e-53
Identities = 98/141 (69%), Positives = 122/141 (86%)
Frame = +3
Query: 3 AVSANLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHV 62
AV+ +LPR + FLL +FVLCSSGAKVV IDVHAAK LIQTG IYLDVRTVEEF+KGHV
Sbjct: 240 AVTVAMLPRW-SVFLLFLFVLCSSGAKVVAIDVHAAKRLIQTGSIYLDVRTVEEFKKGHV 416
Query: 63 DATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGF 122
DA ++NIPY+L+TPKG+VKN +F+K+VSS+C+KED L++GCQSG RS AT++LL++GF
Sbjct: 417 DAVNVLNIPYMLNTPKGKVKNPDFLKEVSSACNKEDHLILGCQSGVRSLYATADLLSEGF 596
Query: 123 KNVHNMGGGYMEWVSNKLPVI 143
KNV +MGGGY++WV NK PVI
Sbjct: 597 KNVKDMGGGYVDWVKNKFPVI 659
>TC229613 weakly similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (40%)
Length = 872
Score = 176 bits (445), Expect = 5e-45
Identities = 80/118 (67%), Positives = 101/118 (84%)
Frame = +2
Query: 25 SSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNL 84
SSG +VVT+DVHA K+LIQT H+YLDVRTVEEF+KGHVDA KIIN+ Y+ +TP+GRVKN
Sbjct: 209 SSGPEVVTVDVHATKDLIQTSHVYLDVRTVEEFQKGHVDAEKIINVAYMFNTPEGRVKNP 388
Query: 85 NFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
F+K+VS +C KED ++VGCQSG RS AT++LL +GFK+V NMGGGY++WV N+LPV
Sbjct: 389 EFLKEVSYACKKEDHIIVGCQSGVRSLYATADLLTEGFKDVSNMGGGYLDWVKNELPV 562
>TC231734 weakly similar to UP|Q7Y234 (Q7Y234) At2g21045, partial (81%)
Length = 647
Score = 120 bits (300), Expect = 3e-28
Identities = 62/108 (57%), Positives = 77/108 (70%), Gaps = 1/108 (0%)
Frame = +1
Query: 30 VVTIDVHAAKNLIQT-GHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVK 88
VVTIDVHAAK+L+ + G+ YLDVR+VEEF K HV+ N+PY+ T GRVKN +FV
Sbjct: 76 VVTIDVHAAKDLLNSSGYRYLDVRSVEEFNKSHVENAH--NVPYVFITEAGRVKNPDFVD 249
Query: 89 QVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWV 136
QV++ C ED L+V C SG RS A+ +LL GFKN+ NMGGGY WV
Sbjct: 250 QVAAICKNEDHLIVACNSGGRSLRASVDLLDSGFKNIVNMGGGYSAWV 393
>TC204706 similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (61%)
Length = 921
Score = 108 bits (271), Expect = 7e-25
Identities = 55/110 (50%), Positives = 69/110 (62%)
Frame = +3
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A L+ GH YLDVRT EEF GH A INIPY+ G KN NF+++VS
Sbjct: 315 SVPVRVAYELLLAGHRYLDVRTPEEFNAGH--APGAINIPYMFRVGSGMTKNSNFIREVS 488
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLP 141
S+ KED ++VGC+ GKRS A S+LLA GF + +M GGY W N LP
Sbjct: 489 SNFRKEDEIIVGCELGKRSMMAASDLLAAGFTGLTDMAGGYAAWTQNGLP 638
>TC204705 similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (61%)
Length = 851
Score = 108 bits (269), Expect = 1e-24
Identities = 54/110 (49%), Positives = 69/110 (62%)
Frame = +3
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A L+ GH YLDVRT EEF+ GH A INIPY+ G KN NF+++VS
Sbjct: 303 SVPVRVAYELLLAGHRYLDVRTPEEFDAGH--APGAINIPYMFRVGSGMTKNSNFIREVS 476
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLP 141
S K+D ++VGC+ GKRS A S+LLA GF + +M GGY W N LP
Sbjct: 477 SQFRKDDEIIVGCELGKRSMMAASDLLAAGFTGLTDMAGGYAAWTQNGLP 626
>TC228316 similar to UP|Q94A65 (Q94A65) AT4g27700/T29A15_190, partial (75%)
Length = 1012
Score = 46.6 bits (109), Expect = 4e-06
Identities = 37/149 (24%), Positives = 63/149 (41%), Gaps = 32/149 (21%)
Frame = +2
Query: 29 KVVTIDVHAAKNLI-QTGHIYLDVRTVEEFEKGHVDATKIINIPYLL------DTPK--- 78
+V +++V A L + + LDVR EF++ H + I L+ D +
Sbjct: 371 RVRSVEVKEALRLQKENSFVLLDVRPEAEFKEAHPPGAINVQIYRLIKEWTAWDIARRAA 550
Query: 79 --------GRVKNLNFVKQVSSSCDKEDCLVVGCQSG--------------KRSFSATSE 116
G +N F+K V + DK+ ++V C SG RS A
Sbjct: 551 FLFFGIFSGTEENPEFIKNVEAKIDKDAKIIVACTSGGTLRPSQNLPEGQQSRSLIAAYL 730
Query: 117 LLADGFKNVHNMGGGYMEWVSNKLPVIQQ 145
L+ +G+ NV ++ GG +W +LP + +
Sbjct: 731 LVLNGYTNVFHLEGGLYKWFKEELPTVSE 817
>TC227788 weakly similar to UP|Q8LBR6 (Q8LBR6) Rhodanese-like family protein,
partial (79%)
Length = 887
Score = 43.5 bits (101), Expect = 4e-05
Identities = 38/146 (26%), Positives = 62/146 (42%), Gaps = 24/146 (16%)
Frame = +1
Query: 23 LCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIP-YLLDTPKGRV 81
L SG + A+ + G + LDVR E EK V + +++P ++ DT +
Sbjct: 196 LIESGTVRPILPKDASTAINSEGFVLLDVRPTWEREKARVAGS--LHVPMFVEDTDNSPI 369
Query: 82 K----------------------NLNFVKQVSSSCD-KEDCLVVGCQSGKRSFSATSELL 118
N F+ QV ++ KE L+V C G RS +A S+L
Sbjct: 370 TLLKKWVHFGYIGLWTGQYLTTLNSEFLSQVENAIPGKEAKLLVACGEGLRSMTAASKLY 549
Query: 119 ADGFKNVHNMGGGYMEWVSNKLPVIQ 144
G+KN+ + GG+ +N P ++
Sbjct: 550 NGGYKNLGWLAGGFNRSKNNDFPAVE 627
>TC215910 similar to UP|O48529 (O48529) Rhodanese-like family protein,
partial (82%)
Length = 1154
Score = 42.4 bits (98), Expect = 8e-05
Identities = 34/122 (27%), Positives = 54/122 (43%), Gaps = 24/122 (19%)
Frame = +3
Query: 28 AKVVTIDVHAAKNLIQT-GHIYLDVRTVEEFEKGHVDATKIINIPYLL---DTPKGRV-- 81
A+V ++ AK L++ G+ LDVR +F + H+ + ++P + D G +
Sbjct: 174 AEVKYVNAEKAKELVEADGYTVLDVRDKTQFVRAHIKSCS--HVPLFVENKDNDPGTIIK 347
Query: 82 ------------------KNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFK 123
N FV+ V S E L+V CQ G RS +A S+L GF+
Sbjct: 348 RQLHNNFSGLFFGLPFTKPNPEFVQSVKSQFPPESKLLVVCQEGLRSAAAASKLEEAGFE 527
Query: 124 NV 125
N+
Sbjct: 528 NI 533
>TC211315 similar to UP|Q9LMU3 (Q9LMU3) F2H15.8 protein, partial (33%)
Length = 898
Score = 39.7 bits (91), Expect = 5e-04
Identities = 28/105 (26%), Positives = 49/105 (46%), Gaps = 1/105 (0%)
Frame = +3
Query: 38 AKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKE 97
AK+ +I LDVR E++ GH + ++ +T G + S+ DKE
Sbjct: 579 AKDNSNRNYILLDVRNGYEWDIGHFRGAQRPSVDCFRNTSFGLSREEITASDPLSNVDKE 758
Query: 98 DC-LVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLP 141
+++ C G R +S L GF+N++ + GG ++ N+ P
Sbjct: 759 KANILMYCTGGIRCDVYSSILRQQGFQNLYTLKGGVSHYLKNEGP 893
>TC215911 similar to UP|Q6H444 (Q6H444) Rhodanese family protein-like,
partial (48%)
Length = 734
Score = 37.0 bits (84), Expect = 0.003
Identities = 20/43 (46%), Positives = 25/43 (57%)
Frame = +1
Query: 83 NLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNV 125
N FV V S E L+V CQ G RS +A S+L A GF+N+
Sbjct: 16 NPEFVPSVKSQFPPESKLLVVCQEGLRSAAAASKLEAAGFENI 144
>TC228403 similar to UP|Q6ZI49 (Q6ZI49) Rhodanese-like domain-containing
protein-like, partial (63%)
Length = 1101
Score = 29.6 bits (65), Expect = 0.53
Identities = 15/40 (37%), Positives = 22/40 (54%)
Frame = +1
Query: 86 FVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNV 125
F+ +V K+ L+V CQ G RS +A L G+KN+
Sbjct: 268 FLAKVEEKFPKDAELIVVCQKGLRSLAACELLYNAGYKNL 387
>TC222565 similar to UP|Q7XJN9 (Q7XJN9) At2g40760 protein, partial (34%)
Length = 722
Score = 29.3 bits (64), Expect = 0.69
Identities = 14/33 (42%), Positives = 21/33 (63%)
Frame = +1
Query: 104 CQSGKRSFSATSELLADGFKNVHNMGGGYMEWV 136
C G R ATS LL+ GFK V ++ GG ++++
Sbjct: 268 CT*GIRCEKATSLLLSKGFKEVFHLEGGILKYL 366
>CA820219 weakly similar to GP|20197120|gb| expressed protein {Arabidopsis
thaliana}, partial (7%)
Length = 422
Score = 28.5 bits (62), Expect = 1.2
Identities = 15/25 (60%), Positives = 16/25 (64%), Gaps = 2/25 (8%)
Frame = -1
Query: 52 RTVEEFEKGHVDATK--IINIPYLL 74
RT EEF GHVDA K IN Y+L
Sbjct: 188 RTAEEFSSGHVDARKHEKINASYVL 114
>CA953362
Length = 426
Score = 27.7 bits (60), Expect = 2.0
Identities = 13/46 (28%), Positives = 23/46 (49%)
Frame = +3
Query: 100 LVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPVIQQ 145
L++G + K+ +S GFK+ +M W+SN + +QQ
Sbjct: 249 LIIGSKRVKKQIEESS-----GFKSYVKQDSSFMNWISNMVKGLQQ 371
>TC204363 similar to UP|O22403 (O22403) ADL2 protein, partial (15%)
Length = 1368
Score = 27.7 bits (60), Expect = 2.0
Identities = 12/27 (44%), Positives = 19/27 (69%)
Frame = -3
Query: 110 SFSATSELLADGFKNVHNMGGGYMEWV 136
S ++ SELL+DG K +GGG ++W+
Sbjct: 358 SIASVSELLSDGLK----IGGGSLKWI 290
>TC204460 homologue to UP|B2_DAUCA (P37707) B2 protein, partial (71%)
Length = 942
Score = 26.9 bits (58), Expect = 3.4
Identities = 21/49 (42%), Positives = 24/49 (48%), Gaps = 7/49 (14%)
Frame = -1
Query: 8 LLPRCLAFFLLLVF----VLCSSGAKVVTIDVHAAKNLIQ---TGHIYL 49
LLP + FFLL F +LC K VT V KN+ Q GHI L
Sbjct: 576 LLPL*VMFFLLYYFAIYLLLCRQSLKSVTEGVLFCKNIQQRQRLGHIQL 430
>TC234040
Length = 850
Score = 26.9 bits (58), Expect = 3.4
Identities = 20/53 (37%), Positives = 27/53 (50%), Gaps = 4/53 (7%)
Frame = -3
Query: 2 AAVSANLLPRCLAFFLLLVFVLCSSGAKVVT----IDVHAAKNLIQTGHIYLD 50
A++ A P C AFFLL + LCS ++T I V NL TG I ++
Sbjct: 623 ASILAVETPICNAFFLLCLLFLCSGLQWLLTETNIIRVVNVYNLPVTGSILME 465
>CO979705
Length = 709
Score = 26.6 bits (57), Expect = 4.5
Identities = 14/42 (33%), Positives = 22/42 (52%)
Frame = +3
Query: 13 LAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTV 54
L LLL+ +LC SGA + + AA ++ G + + TV
Sbjct: 108 LLLLLLLLLLLCCSGAPPIMEVIAAANAAVKFGSVAMAAPTV 233
>BQ252470
Length = 256
Score = 26.6 bits (57), Expect = 4.5
Identities = 12/23 (52%), Positives = 14/23 (60%)
Frame = +2
Query: 12 CLAFFLLLVFVLCSSGAKVVTID 34
C FFLLLVF+ SG V +D
Sbjct: 104 CYFFFLLLVFLFADSGTGVRGVD 172
>TC205252 homologue to UP|Q9ZP50 (Q9ZP50) FtsH-like protein Pftf precursor,
partial (61%)
Length = 1482
Score = 26.2 bits (56), Expect = 5.9
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 9 LPRCLAFFLLLVFVLCSSGAK 29
L RCL +LLVFV+CS K
Sbjct: 80 LLRCLLVLVLLVFVICSRRRK 142
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,906,721
Number of Sequences: 63676
Number of extensions: 90659
Number of successful extensions: 487
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 481
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 483
length of query: 145
length of database: 12,639,632
effective HSP length: 88
effective length of query: 57
effective length of database: 7,036,144
effective search space: 401060208
effective search space used: 401060208
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC147013.5


