

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147013.12 - phase: 0
(687 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
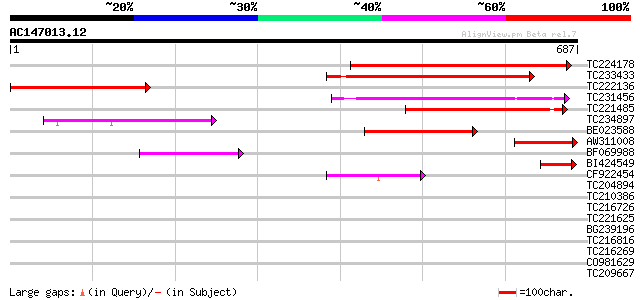
Score E
Sequences producing significant alignments: (bits) Value
TC224178 406 e-113
TC233433 402 e-112
TC222136 similar to UP|HMN1_DROME (P22807) Homeobox protein slou... 258 8e-69
TC231456 similar to UP|Q9U1G9 (Q9U1G9) Gly-rich protein, partial... 181 1e-45
TC221485 162 4e-40
TC234897 weakly similar to UP|TIR3_YEAST (P40552) Cell wall prot... 142 4e-34
BE023588 125 6e-29
AW311008 115 5e-26
BF069988 67 2e-11
BI424549 58 1e-08
CF922454 43 5e-04
TC204894 similar to UP|Q94KA9 (Q94KA9) Importin alpha 2, partial... 39 0.008
TC210386 similar to UP|Q84TU4 (Q84TU4) Arm repeat-containing pro... 38 0.013
TC216726 similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing pro... 37 0.029
TC221625 similar to UP|O82783 (O82783) Importin alpha 2 subunit ... 37 0.038
BG239196 35 0.085
TC216816 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like pro... 35 0.14
TC216269 similar to UP|E6_GOSHI (Q01197) Protein E6, partial (17%) 34 0.19
CO981629 33 0.55
TC209667 similar to GB|BAC22265.1|24414015|AP003803 arm repeat c... 33 0.55
>TC224178
Length = 821
Score = 406 bits (1043), Expect = e-113
Identities = 212/267 (79%), Positives = 233/267 (86%)
Frame = +3
Query: 414 QRNYSHSGINMKGRESEDAETKASMKEMAARALWHLAKGNVAICRSITESRALLCFSVLL 473
+++YS+SGINMKGRE ED + KA MK MAARAL LAKGN AICRSITESRALLC ++LL
Sbjct: 12 EQSYSYSGINMKGREIEDPDNKAYMKAMAARALRQLAKGNAAICRSITESRALLCLAILL 191
Query: 474 EKGPEAVQYNSAMALMEITAVAEKDAELRKSAFKPNSPACKAVVDQVLKIIEKADSDLLI 533
EKG E V YNSA+A+ EITAVAEKDAELR+SAFKPNSPACKAVVDQVLKIIEK D LLI
Sbjct: 192 EKGTEDVMYNSALAVKEITAVAEKDAELRRSAFKPNSPACKAVVDQVLKIIEKEDRKLLI 371
Query: 534 PCVKAIGNLARTFKATETRMIGPLVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNA 593
PCVKAIGNLARTF+ATETR+IGPLV+LLDEREAEVSREA+I+L K A SENYLH+DHS A
Sbjct: 372 PCVKAIGNLARTFRATETRIIGPLVRLLDEREAEVSREAAISLTKLACSENYLHLDHSKA 551
Query: 594 IISAGGAKHLIQLVYFGEQMVQIPALVLLSYIALHVPDSEELALAEVLGVLEWASKQSFM 653
IISA GAKHL+QLVY GEQ VQI ALVLLSYIALHVPDSEELA AEVLGVLEWASKQ +
Sbjct: 552 IISASGAKHLVQLVYLGEQTVQISALVLLSYIALHVPDSEELARAEVLGVLEWASKQPNL 731
Query: 654 QHDETLEELLQEAKSRLELYQSRGSRG 680
D+ LE LLQ++K RL YQSRGSRG
Sbjct: 732 TQDQNLEALLQDSKGRLGAYQSRGSRG 812
>TC233433
Length = 754
Score = 402 bits (1032), Expect = e-112
Identities = 210/252 (83%), Positives = 224/252 (88%), Gaps = 1/252 (0%)
Frame = +2
Query: 385 NSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETKASMKEMAAR 444
N + P NG+GN GN Q + NHQ NYSHSGINMKGR+ ED +TKA+MKEMAAR
Sbjct: 11 NESNPKTNGVGNDAKQGN----QDQNQNHQPNYSHSGINMKGRDHEDPKTKANMKEMAAR 178
Query: 445 ALWHLAKGNVAICRSITESRALLCFSVLLEKGPEAVQYNSAMALMEITAVAEKDAELRKS 504
ALW LAKGN ICRSITESRALLCF+VLLEKG EAVQYNSAMA+MEITAVAEKDAELRKS
Sbjct: 179 ALWQLAKGNSPICRSITESRALLCFAVLLEKGTEAVQYNSAMAVMEITAVAEKDAELRKS 358
Query: 505 AFKPNSPACKAVVDQVLKIIEKADSDLLIPCVKAIGNLARTFKATETRMIGPLVKLLDER 564
AFKPNSPACKAVVDQV+KIIEKADS+LLIPC+K IGNLARTFKATETRMIGPLVKLLDER
Sbjct: 359 AFKPNSPACKAVVDQVVKIIEKADSELLIPCIKTIGNLARTFKATETRMIGPLVKLLDER 538
Query: 565 EAEVSREASIALRKFAGSENYLHVDHSNAIISAGGAKHLIQLVYF-GEQMVQIPALVLLS 623
EAEVSREASIAL KFA +ENYLHVDHS AII AGGAKHLIQLVYF GE+MVQIPALVLLS
Sbjct: 539 EAEVSREASIALTKFACTENYLHVDHSKAIIIAGGAKHLIQLVYFGGEEMVQIPALVLLS 718
Query: 624 YIALHVPDSEEL 635
YIA+HVPDSEEL
Sbjct: 719 YIAMHVPDSEEL 754
>TC222136 similar to UP|HMN1_DROME (P22807) Homeobox protein slou (S59/2)
(Slouch protein) (Homeobox protein NK-1), partial (3%)
Length = 562
Score = 258 bits (658), Expect = 8e-69
Identities = 139/172 (80%), Positives = 150/172 (86%), Gaps = 2/172 (1%)
Frame = +2
Query: 1 MGDIVKQILAKPIQLADQVTKAADEASS-FKQECSELKSKTEKLATLLRQAARASSDLYE 59
M DIVKQ+LAKPIQLADQV KAA+EASS FKQEC ELKSK +KLA LLR AARASSDLYE
Sbjct: 47 MADIVKQLLAKPIQLADQVAKAAEEASSSFKQECLELKSKADKLAALLRLAARASSDLYE 226
Query: 60 RPTKRIIEETEQVLDKALSLVLKCRANGLMKRVFTIIPAAAFRKTSSHLENSIGDVSWLL 119
RPT+RII +TE VLDKALSL LKCRANGLMKRVF+IIP AAFRK SS LENSIGDVSWLL
Sbjct: 227 RPTRRIIADTELVLDKALSLTLKCRANGLMKRVFSIIPTAAFRKMSSQLENSIGDVSWLL 406
Query: 120 RVSAPADDRGG-EYLGLPPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASL 170
RVS PA++R EYLGLPPIAANEPIL IWEQ+A+L TGS + RSDAAASL
Sbjct: 407 RVSTPAEERADTEYLGLPPIAANEPILGLIWEQVAVLHTGSLDDRSDAAASL 562
>TC231456 similar to UP|Q9U1G9 (Q9U1G9) Gly-rich protein, partial (6%)
Length = 971
Score = 181 bits (458), Expect = 1e-45
Identities = 110/290 (37%), Positives = 167/290 (56%), Gaps = 1/290 (0%)
Frame = +1
Query: 390 NGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETKASMKEMAARALWHL 449
N + +G N +G+ G SG + + RE E E + +K A+ALW L
Sbjct: 7 NLSNVGEANSDGSSRG--------------SGQHRRDREVESPELRNEVKVSCAKALWKL 144
Query: 450 AKGNVAICRSITESRALLCFSVLLEKGPEAVQYNSAMALMEITAVAEKDAELRKSAFKPN 509
+ G ++ CR ITE++ LLC + ++E +Q N MA+MEI AVAE +A+LR++AFK
Sbjct: 145 SNGCLSSCRKITETKGLLCLAKIIESESGELQLNCFMAVMEIAAVAESNADLRRAAFKRT 324
Query: 510 SPACKAVVDQVLKII-EKADSDLLIPCVKAIGNLARTFKATETRMIGPLVKLLDEREAEV 568
+P KAV+DQ+L+++ E++D L IP +KAIG+LAR F ++IGPLV L R+ +V
Sbjct: 325 APVAKAVLDQLLRVVQEESDPALRIPAIKAIGSLARNFSGKVPQVIGPLVAQLGNRDVDV 504
Query: 569 SREASIALRKFAGSENYLHVDHSNAIISAGGAKHLIQLVYFGEQMVQIPALVLLSYIALH 628
+ EA+IAL KF +NY VDHS AI+ G L+ L+ ++ Q+ L LL Y+AL+
Sbjct: 505 ASEAAIALGKFVCPDNYNCVDHSKAILELDGIPKLMSLLQINDRQ-QVHGLKLLCYLALN 681
Query: 629 VPDSEELALAEVLGVLEWASKQSFMQHDETLEELLQEAKSRLELYQSRGS 678
V +S L L LE ++ QH + L++L +A + +R S
Sbjct: 682 VGNSRVLEQERALNTLERFARPVQAQHPD-LKDLFAKALHQPHSLSARSS 828
>TC221485
Length = 773
Score = 162 bits (411), Expect = 4e-40
Identities = 95/197 (48%), Positives = 124/197 (62%), Gaps = 1/197 (0%)
Frame = +2
Query: 480 VQYNSAMALMEITAVAEKDAELRKSAFKPNSPACKAVVDQVLKIIEKADSDLL-IPCVKA 538
+Q N M +MEITA AE +A+LR++AFK NSP KAVV+Q+L+II++ DS L IP +KA
Sbjct: 5 LQLNCLMTIMEITAAAESNADLRRAAFKTNSPPAKAVVEQLLRIIKEVDSPALQIPAMKA 184
Query: 539 IGNLARTFKATETRMIGPLVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNAIISAG 598
IG+LARTF ETR+I PLV + R EV+ EA AL KFA +NYLH++HS II
Sbjct: 185 IGSLARTFPVRETRVIEPLVTQMGNRNTEVADEAVAALTKFASPDNYLHIEHSKTIIEFN 364
Query: 599 GAKHLIQLVYFGEQMVQIPALVLLSYIALHVPDSEELALAEVLGVLEWASKQSFMQHDET 658
G L++L+ E L LL Y+ALH +SE L A VL VLE A + QH
Sbjct: 365 GIPALMRLLRSNEVTQMHRGLTLLCYLALHAGNSESLEQARVLTVLEGADRTVLPQH--- 535
Query: 659 LEELLQEAKSRLELYQS 675
++EL+ A L LY +
Sbjct: 536 IKELVSRAIIHLNLYHA 586
>TC234897 weakly similar to UP|TIR3_YEAST (P40552) Cell wall protein TIR3
precursor, partial (12%)
Length = 698
Score = 142 bits (359), Expect = 4e-34
Identities = 85/221 (38%), Positives = 126/221 (56%), Gaps = 11/221 (4%)
Frame = +3
Query: 41 EKLATLLRQAARASSD-----LYERPTKRIIEETEQVLDKALSLVLKCRANGLMKRVFTI 95
+ L TL+R A S+ LYERP +R+ E + LD+AL+LV KC+ +++RV +I
Sbjct: 30 QMLRTLVRFATATSTSSVAPPLYERPIRRVAAEASKNLDRALALVRKCKRRSILRRVVSI 209
Query: 96 IPAAAFRKTSSHLENSIGDVSWLLRV-----SAPADDRGGEYLGLPPIAANEPILCFIWE 150
+ AA FRK +H++ S GD+ WLL + + GG L LPPIA+N+PIL ++W
Sbjct: 210 VSAADFRKVLTHIDASTGDMMWLLSILDADGAGDGGGGGGIVLSLPPIASNDPILSWVWS 389
Query: 151 QIAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKAD-GQENAA 209
IA + G R +AA L S A+ + R K+I+EE GV PLLKL+KEG + Q AA
Sbjct: 390 FIASIQMGQLNDRIEAANELASFAQDNARNKKIIVEECGVPPLLKLLKEGTSPLAQIAAA 569
Query: 210 RAIGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVA 250
+ L D + V ++ ++L + PM+VQ + A
Sbjct: 570 TTLCHLANDLDRVRVIVSEHGVPAVVQVLSDSPMRVQTLAA 692
>BE023588
Length = 424
Score = 125 bits (314), Expect = 6e-29
Identities = 64/139 (46%), Positives = 95/139 (68%), Gaps = 1/139 (0%)
Frame = +3
Query: 430 EDAETKASMKEMAARALWHLAKGNVAICRSITESRALLCFSVLLEKGPEAVQYNSAMALM 489
E E + +K A+ALW L+KG ++ CR ITE++ LLC + ++E +Q N MA+M
Sbjct: 6 ESPELRNEVKISCAKALWKLSKGCLSSCRKITETKGLLCLAKIIESESGELQLNCLMAVM 185
Query: 490 EITAVAEKDAELRKSAFKPNSPACKAVVDQVLKII-EKADSDLLIPCVKAIGNLARTFKA 548
EI AVAE +A+LR++AFK +PA KAV+DQ+L+++ E++D L IP +KAIG+LAR F
Sbjct: 186 EIAAVAESNADLRRAAFKRTAPAAKAVLDQLLRVVQEESDPALRIPAIKAIGSLARNFSG 365
Query: 549 TETRMIGPLVKLLDEREAE 567
++IGPLV L R+ +
Sbjct: 366 KVPQVIGPLVAQLGNRDVD 422
>AW311008
Length = 378
Score = 115 bits (289), Expect = 5e-26
Identities = 63/76 (82%), Positives = 65/76 (84%)
Frame = -3
Query: 612 QMVQIPALVLLSYIALHVPDSEELALAEVLGVLEWASKQSFMQHDETLEELLQEAKSRLE 671
Q VQI ALVLLSYIALHVPDSEELA AEVLGVLEWASKQ + DETLE LLQE+K RLE
Sbjct: 376 QTVQISALVLLSYIALHVPDSEELARAEVLGVLEWASKQPNVTQDETLEALLQESKGRLE 197
Query: 672 LYQSRGSRGFHHKLHQ 687
LYQSRGSRGF KLHQ
Sbjct: 196 LYQSRGSRGF-QKLHQ 152
>BF069988
Length = 400
Score = 67.4 bits (163), Expect = 2e-11
Identities = 42/128 (32%), Positives = 69/128 (53%), Gaps = 2/128 (1%)
Frame = +3
Query: 158 GSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAA--RAIGLL 215
G + R++AA L SLAR +DR +I++EGGV PLLKL+KE + + AA + +
Sbjct: 15 GQPKDRAEAATELGSLARDNDRTKFIILDEGGVMPLLKLLKEASSPAAQVAAANALVNIT 194
Query: 216 GRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHNIIRL 275
V ++ + ++L + PM+V+ VA VS +A + +E F + N+ R
Sbjct: 195 TNQDRVVTFIVESHAVPIIVQVLGDSPMRVRVSVANLVSAMAEQHELAREEFVRANVARP 374
Query: 276 LVGHLAFE 283
LV L+ +
Sbjct: 375 LVSLLSMD 398
>BI424549
Length = 348
Score = 58.2 bits (139), Expect = 1e-08
Identities = 27/43 (62%), Positives = 36/43 (82%)
Frame = -1
Query: 644 LEWASKQSFMQHDETLEELLQEAKSRLELYQSRGSRGFHHKLH 686
+EWASKQS + +D+ +E LL E+K++L+LYQSRG RGF HKLH
Sbjct: 348 IEWASKQSSIANDQAIEALLLESKTKLDLYQSRGPRGF-HKLH 223
>CF922454
Length = 785
Score = 42.7 bits (99), Expect = 5e-04
Identities = 34/128 (26%), Positives = 52/128 (40%), Gaps = 8/128 (6%)
Frame = +2
Query: 385 NSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETKASMKEMAAR 444
N N NGNG GNGNG GND ++ + K +E E E + K+
Sbjct: 197 NGNNGNGNGNGNGNGKGNDKEEKEREKREKEKEKREKKEKKEKEKEKKEKEKQAKKQQGE 376
Query: 445 A-----LWHLAKGNV-AICRSIT--ESRALLCFSVLLEKGPEAVQYNSAMALMEITAVAE 496
A L L G CR T E + ++C S + P+ + A + ++ E
Sbjct: 377 ATNYDKLSPLPTGQERGFCRKNTACEFKTIVCPSECAYRKPKKNKKQKACFIDCSSSTCE 556
Query: 497 KDAELRKS 504
++RK+
Sbjct: 557 ATCKVRKA 580
Score = 35.4 bits (80), Expect = 0.085
Identities = 22/57 (38%), Positives = 26/57 (45%), Gaps = 1/57 (1%)
Frame = +2
Query: 390 NGNGL-GNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETKASMKEMAARA 445
NGNG GNGNGNGN GK GN + K + E E + KE +A
Sbjct: 191 NGNGNNGNGNGNGNGNGK-GNDKEEKEREKREKEKEKREKKEKKEKEKEKKEKEKQA 358
Score = 34.3 bits (77), Expect = 0.19
Identities = 19/50 (38%), Positives = 23/50 (46%)
Frame = +2
Query: 395 GNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETKASMKEMAAR 444
G GNGNGN+G GN N N KG + E+ E + KE R
Sbjct: 182 GQGNGNGNNGNGNGNGNG----------NGKGNDKEEKEREKREKEKEKR 301
Score = 31.6 bits (70), Expect = 1.2
Identities = 15/43 (34%), Positives = 19/43 (43%)
Frame = +2
Query: 393 GLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETK 435
G GNGNGN +G GN N + + +E E E K
Sbjct: 182 GQGNGNGNNGNGNGNGNGNGKGNDKEEKEREKREKEKEKREKK 310
>TC204894 similar to UP|Q94KA9 (Q94KA9) Importin alpha 2, partial (95%)
Length = 2332
Score = 38.9 bits (89), Expect = 0.008
Identities = 28/113 (24%), Positives = 55/113 (47%), Gaps = 2/113 (1%)
Frame = +2
Query: 184 IIEEGGVGPLLK-LIKEGKADGQENAARAI-GLLGRDAESVEHMIHVGVCSVFAKILKEG 241
+I+ G V ++ L++E Q AA A+ + +E+ + +I G +F K+L
Sbjct: 497 VIQTGVVSRFVEFLMREDFPQLQFEAAWALTNIASGTSENTKVVIDHGAVPIFVKLLASP 676
Query: 242 PMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLVGHLAFETVEEHSKYAIV 294
V+ WA+ +A + P+C++L H + L+ L EH+K +++
Sbjct: 677 SDDVREQAVWALGNVAGDSPRCRDLVLSHGALLPLLAQL-----NEHAKLSML 820
>TC210386 similar to UP|Q84TU4 (Q84TU4) Arm repeat-containing protein,
partial (13%)
Length = 653
Score = 38.1 bits (87), Expect = 0.013
Identities = 18/57 (31%), Positives = 34/57 (59%)
Frame = +1
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENA 208
+ ++ +GS + +AAA+L+ L + RY ++++EG V PL+ L + G +E A
Sbjct: 76 VEVIESGSARGKENAAAALLHLCSDNHRYLNMVLQEGAVPPLVALSQSGTPRAKEKA 246
Score = 32.3 bits (72), Expect = 0.72
Identities = 19/59 (32%), Positives = 33/59 (55%)
Frame = +1
Query: 168 ASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAESVEHMI 226
A+L ++ G G ++GG+ L+++I+ G A G+ENAA A+ L D +M+
Sbjct: 10 ANLATIPEGKTAIG----QQGGIPVLVEVIESGSARGKENAAAALLHLCSDNHRYLNMV 174
>TC216726 similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing protein
homolog, partial (35%)
Length = 1115
Score = 37.0 bits (84), Expect = 0.029
Identities = 22/74 (29%), Positives = 36/74 (47%)
Frame = +3
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
+ + GS + +AAA LV L G +Y E G +GPLL+L + G G+ A +
Sbjct: 477 VEFIGNGSPRNKENAAAVLVHLCSGDQQYLAQAQELGVMGPLLELAQNGTDRGKRKAGQL 656
Query: 212 IGLLGRDAESVEHM 225
+ + R E + +
Sbjct: 657 LERMSRLVEQQQEV 698
>TC221625 similar to UP|O82783 (O82783) Importin alpha 2 subunit (NLS
receptor), partial (62%)
Length = 988
Score = 36.6 bits (83), Expect = 0.038
Identities = 25/100 (25%), Positives = 48/100 (48%), Gaps = 1/100 (1%)
Frame = +3
Query: 196 LIKEGKADGQENAARAI-GLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVS 254
L++E Q AA A+ + +E+ + +I G +F K+L V+ WA+
Sbjct: 174 LMREDFPQLQFEAAWALTNIASGTSENTKVIIDHGAVPIFVKLLGSPSDDVREQAVWALG 353
Query: 255 ELAANYPKCQELFAQHNIIRLLVGHLAFETVEEHSKYAIV 294
+A + P+C++L H + L+ L EH+K +++
Sbjct: 354 NVAGDSPRCRDLVLGHGALLPLLAQL-----NEHAKLSML 458
>BG239196
Length = 405
Score = 35.4 bits (80), Expect = 0.085
Identities = 20/81 (24%), Positives = 40/81 (48%), Gaps = 2/81 (2%)
Frame = +3
Query: 184 IIEEGGVGPLLKLIKEGKADGQENAARAIGLL--GRDAESVEHMIHVGVCSVFAKILKEG 241
+IE GG+ ++ ++K GK + +ENA A+ + ES ++ G+ + L G
Sbjct: 9 LIELGGLDAIISILKTGKMEAKENALTALFRFTDPTNIESQRDLVKRGIYPLLVDFLNTG 188
Query: 242 PMKVQGVVAWAVSELAANYPK 262
+ + A + +L+ + PK
Sbjct: 189 SVTAKARAAAFIGDLSMSTPK 251
>TC216816 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like protein X,
partial (5%)
Length = 1610
Score = 34.7 bits (78), Expect = 0.14
Identities = 35/132 (26%), Positives = 55/132 (41%)
Frame = +1
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
I ML + +++ +A +L LA+ + I GG+ PLLKL+ Q NAA A
Sbjct: 88 IEMLQSSDVQLKEMSAFALGRLAQDTHNQAG-IAHNGGLMPLLKLLDSKNGSLQHNAAFA 264
Query: 212 IGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHN 271
+ L + ++V I VG + L++G VQ L K H
Sbjct: 265 LYGLADNEDNVSDFIRVG----GIQRLQDGEFIVQATKDCVAKTLKRLEEKIHGRVLNHL 432
Query: 272 IIRLLVGHLAFE 283
+ + V AF+
Sbjct: 433 LYLMRVSEKAFQ 468
>TC216269 similar to UP|E6_GOSHI (Q01197) Protein E6, partial (17%)
Length = 1139
Score = 34.3 bits (77), Expect = 0.19
Identities = 33/128 (25%), Positives = 54/128 (41%), Gaps = 1/128 (0%)
Frame = +1
Query: 288 HSKYAIVSMKANSIHAAVVMASNNNNSSSNLNPKK-GTENEDGVVVGGGNKHGRVSHHPL 346
H+ Y + K + AA + S NNN++ N N + T N + V G + +S
Sbjct: 487 HNSYQNNNQKYYNNDAANGIYSYNNNNNYNANNNRYNTYNNNNAVNGYNGERQGMSDTRF 666
Query: 347 GERPRNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGK 406
E + H V A+++ N N + ++ + N+N N GN NGN N
Sbjct: 667 LEGGKYFHDV---------AAEKYNPTNYGDSSREV--NTNNWYNNRGGNYNGNQNQQEF 813
Query: 407 QGNHNNHQ 414
+ H N +
Sbjct: 814 EDEHENFE 837
>CO981629
Length = 844
Score = 32.7 bits (73), Expect = 0.55
Identities = 27/89 (30%), Positives = 41/89 (45%), Gaps = 18/89 (20%)
Frame = -3
Query: 367 SKQPNEGNEANQNQNILANSNTPNGNGLGNG-------------NGNGN-----DGGKQG 408
+K N ++N N N NSN+ +G NG NG+ N DG K
Sbjct: 719 NKNNNHNAKSNGNSNDYRNSNSVKSSGTSNGIRNSDCAKSNSNVNGHNNSSCKSDGAK-- 546
Query: 409 NHNNHQRNYSHSGINMKGRESEDAETKAS 437
++NN+ RN + N + SEDA++ A+
Sbjct: 545 SNNNYTRNS*GASSNY-SQTSEDAKSNAN 462
Score = 29.6 bits (65), Expect = 4.6
Identities = 30/148 (20%), Positives = 54/148 (36%), Gaps = 5/148 (3%)
Frame = -3
Query: 295 SMKANSIHAAVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLH 354
S K N + + NNN ++ N V G +G + + + N++
Sbjct: 761 SAKNNPRNGDDAYGNKNNNHNAKSNGNSNDYRNSNSVKSSGTSNG-IRNSDCAKSNSNVN 585
Query: 355 RVITSTMAIHAASKQPNE-----GNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGN 409
S+ A N G +N +Q + NGN + +G G + N
Sbjct: 584 GHNNSSCKSDGAKSNNNYTRNS*GASSNYSQTSEDAKSNANGNNNSSCKSDGASGNGKSN 405
Query: 410 HNNHQRNYSHSGINMKGRESEDAETKAS 437
+N RN N G +++++ K+S
Sbjct: 404 YN---RNSDGPKSNAIGNDNQNSGAKSS 330
>TC209667 similar to GB|BAC22265.1|24414015|AP003803 arm repeat containing
protein homolog-like {Oryza sativa (japonica
cultivar-group);} , partial (41%)
Length = 787
Score = 32.7 bits (73), Expect = 0.55
Identities = 19/43 (44%), Positives = 27/43 (62%)
Frame = +2
Query: 166 AAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENA 208
A L SLA G + + I+EEGG+G LL+ I++G G+E A
Sbjct: 338 AMVVLNSLA-GIEEGKEAIVEEGGIGALLEAIEDGSVKGKEFA 463
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.131 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,260,187
Number of Sequences: 63676
Number of extensions: 332565
Number of successful extensions: 1857
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 1681
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1800
length of query: 687
length of database: 12,639,632
effective HSP length: 104
effective length of query: 583
effective length of database: 6,017,328
effective search space: 3508102224
effective search space used: 3508102224
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147013.12


