

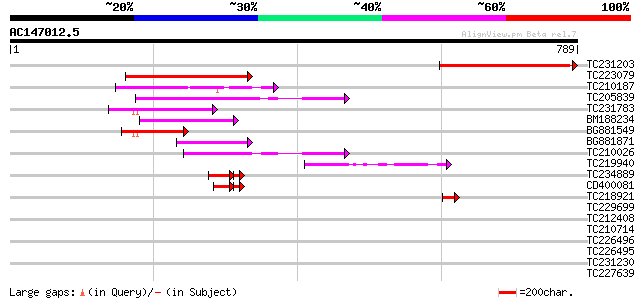
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147012.5 + phase: 0
(789 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC231203 335 6e-92
TC223079 homologue to UP|Q6YUI7 (Q6YUI7) RNA helicase-like, part... 151 1e-36
TC210187 weakly similar to UP|DHX9_HUMAN (Q08211) ATP-dependent ... 136 3e-32
TC205839 homologue to UP|Q9LZQ9 (Q9LZQ9) ATP-dependent RNA helic... 130 3e-30
TC231783 homologue to UP|Q9LZQ9 (Q9LZQ9) ATP-dependent RNA helic... 111 1e-24
BM188234 97 4e-20
BG881549 homologue to PIR|T48023|T48 ATP-dependent RNA helicase-... 80 3e-15
BG881871 74 2e-13
TC210026 homologue to UP|Q9LXT8 (Q9LXT8) Pre-mRNA splicing facto... 67 2e-11
TC219940 similar to UP|O49516 (O49516) RNA helicase - like prote... 50 3e-06
TC234889 weakly similar to UP|Q9SZ53 (Q9SZ53) Protein phosphatas... 46 2e-05
CD400081 45 4e-05
TC218921 weakly similar to GB|AAP37732.1|30725420|BT008373 At1g1... 44 2e-04
TC229699 similar to UP|Q93Y39 (Q93Y39) ATP-dependent RNA helicas... 40 0.005
TC212408 similar to UP|Q867T7 (Q867T7) P67-like superoxide-gener... 38 0.020
TC210714 weakly similar to UP|Q9VIZ3 (Q9VIZ3) CG10689-PA (LD2569... 36 0.058
TC226496 homologue to UP|Q93VJ8 (Q93VJ8) AT5g11200/F2I11_90 (AT5... 34 0.22
TC226495 homologue to UP|Q93VJ8 (Q93VJ8) AT5g11200/F2I11_90 (AT5... 34 0.29
TC231230 weakly similar to GB|CAA65563.1|1403551|SCLACHXVI P2570... 33 0.37
TC227639 similar to UP|E74B_DROME (P11536) Ecdysone-induced prot... 33 0.49
>TC231203
Length = 1136
Score = 335 bits (858), Expect = 6e-92
Identities = 160/191 (83%), Positives = 174/191 (90%)
Frame = +1
Query: 599 MHGVTNVDPTWLVENAKSSCIFSPPLTDPRPFYDAQADQVKCWVIPTFGRFCWELPKHSI 658
MHGVT+V+P WLVE+AKSSCIFSPPL DPRP+YDAQ DQVKCWVIPTFGRFCWELPKHS+
Sbjct: 1 MHGVTSVEPAWLVEHAKSSCIFSPPLMDPRPYYDAQTDQVKCWVIPTFGRFCWELPKHSL 180
Query: 659 PISNVEHRVQVFAYALLEGQVCTCLKSVRKYMSAPPETILRREALGQKRVGNLISKLNSR 718
ISN EHRVQVFAYALLEGQVC CLKSVRKYMSA PE+I++REALGQKRVGNL+SKL SR
Sbjct: 181 SISNDEHRVQVFAYALLEGQVCPCLKSVRKYMSAAPESIMKREALGQKRVGNLLSKLKSR 360
Query: 719 LIDSSAMLRIVWKQNPRELFSEILDWFQQGFRKHFEELWLQMLGEVLQETQEQPLHKSLK 778
LIDSSAMLR+VWK+NPRELFSEILDWFQQ F KHFEELWLQM+ EVL E QE+PLHKS K
Sbjct: 361 LIDSSAMLRMVWKENPRELFSEILDWFQQSFHKHFEELWLQMVNEVLMEKQERPLHKSSK 540
Query: 779 RKSKVKSKLRR 789
+K KVKSK R
Sbjct: 541 KK-KVKSKSLR 570
>TC223079 homologue to UP|Q6YUI7 (Q6YUI7) RNA helicase-like, partial (19%)
Length = 613
Score = 151 bits (381), Expect = 1e-36
Identities = 84/177 (47%), Positives = 116/177 (65%)
Frame = +2
Query: 162 AAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDSSNGMETYEVK 221
A Q ++F+ EG R VV+ATN+AETSLTI GIKYV+D G K+K+Y+ GME+ V
Sbjct: 5 AELQAKIFEPTPEGARKVVLATNIAETSLTIDGIKYVIDPGFCKMKSYNPRTGMESLLVT 184
Query: 222 WISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQI 281
ISKASA QRAGR+GRT G C+RLY++ + N+ + + E+++ + VVL LKS+ I
Sbjct: 185 PISKASANQRAGRSGRTGPGKCFRLYTAYNYHNDLDDNTVPEIQRTNLANVVLTLKSLGI 364
Query: 282 KKVANFPFPTSLKAASLLEAENCLRALEALDSKDELTLLGKAMALYPLSPRHSRMIL 338
+ NF F A +LL+A L AL AL+ ELT +G+ MA +PL P S+MI+
Sbjct: 365 HDLLNFDFMDPPPAEALLKALELLFALSALNKLGELTKVGRRMAEFPLDPMLSKMIV 535
>TC210187 weakly similar to UP|DHX9_HUMAN (Q08211) ATP-dependent RNA helicase
A (Nuclear DNA helicase II) (NDH II) (DEAH-box protein
9), partial (7%)
Length = 691
Score = 136 bits (343), Expect = 3e-32
Identities = 84/234 (35%), Positives = 127/234 (53%), Gaps = 7/234 (2%)
Frame = +2
Query: 148 PGALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVK 207
P +LPL+ +P Q +F+ +R +V+ATN+AE+S+TI + YV+D G+ K
Sbjct: 2 PSKFLILPLHGSMPTVNQCEIFERPPPNKRKIVLATNIAESSITIDDVVYVIDWGKAKET 181
Query: 208 NYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKV 267
+YD+ N + WISKASA QR GRAGR G CYRLY + P++ AE+ +
Sbjct: 182 SYDALNKLACLLPSWISKASAHQRRGRAGRVQPGVCYRLYPK-LIHDAMPQYQLAEILRT 358
Query: 268 PVHGVVLLLKSMQIKKVANF-------PFPTSLKAASLLEAENCLRALEALDSKDELTLL 320
P+ + L +KS+Q+ V +F P P ++K A L L+ + ALD ++ELT L
Sbjct: 359 PLQELCLHIKSLQLGTVGSFLEKALQPPDPLAVKNAIEL-----LKTIGALDEQEELTPL 523
Query: 321 GKAMALYPLSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSLPNPFVM 374
G+ + PL P +M+L + + L A+ AAAL+ NPFV+
Sbjct: 524 GRHLCNIPLDPNIGKMLL----------MGSIFQCLNPALTIAAALAYRNPFVL 655
>TC205839 homologue to UP|Q9LZQ9 (Q9LZQ9) ATP-dependent RNA helicase-like
protein (At3g62310), partial (49%)
Length = 1344
Score = 130 bits (326), Expect = 3e-30
Identities = 88/298 (29%), Positives = 144/298 (47%)
Frame = +1
Query: 176 ERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRA 235
E +VV+TN+AETSLTI GI YV+D G K K Y+ +E+ V ISKASA QR+GRA
Sbjct: 13 EGKIVVSTNIAETSLTIDGIVYVIDPGFAKQKVYNPRVRVESLLVSPISKASAHQRSGRA 192
Query: 236 GRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTSLKA 295
GRT G C+RLY+ +F+N+ + E+ + + VL LK + I + +F F
Sbjct: 193 GRTQPGKCFRLYTEKSFNNDLQPQTYPEILRSNLANTVLTLKKLGIDDLVHFDFMDPPAP 372
Query: 296 ASLLEAENCLRALEALDSKDELTLLGKAMALYPLSPRHSRMILTVIKNTRYKHIRNSSLL 355
+L+ A L L ALD LT LG+ M+ +PL P+ S+M++ + I
Sbjct: 373 ETLMRALEVLNYLGALDDDGNLTKLGQIMSEFPLDPQMSKMLVVSPEFNCSNEI------ 534
Query: 356 LAYAVAAAAALSLPNPFVMQYEGNDSNKDSETSEKSRMGDNENNIDKTEKTKRKKLKQTS 415
++ +A LS+PN FV + ++ ++ +
Sbjct: 535 ----LSVSAMLSVPNCFV---------------------------------RPREAQKAA 603
Query: 416 KVAREKFRIVSSDALAIAYALQCFEHSQNSVQFCEDNALHFKTMDEMSKLRQQLLRLV 473
A+ + D L + ++ + + +C DN ++ + + +RQQL+R++
Sbjct: 604 DEAKASLGHIDGDHLTLLNVYHAYKQNNDDPSWCYDNFVNHRALKSADSVRQQLVRIM 777
>TC231783 homologue to UP|Q9LZQ9 (Q9LZQ9) ATP-dependent RNA helicase-like
protein (At3g62310), partial (24%)
Length = 529
Score = 111 bits (278), Expect = 1e-24
Identities = 70/161 (43%), Positives = 97/161 (59%), Gaps = 9/161 (5%)
Frame = +2
Query: 138 KIARE--NHDSSPGALFVLPLYAMLPAAAQLRVFDG----VKEGE---RLVVVATNVAET 188
KI +E N G + V+PLY+ LP A Q ++F+ VKEG R +VV+TN+AET
Sbjct: 23 KINKEISNMGDQVGPVKVVPLYSTLPPAMQQKIFEPAPPPVKEGGPPGRKIVVSTNIAET 202
Query: 189 SLTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYS 248
SLTI GI YV+D G K K Y+ +E+ V ISKASA QR+GRAGRT G C+RLY+
Sbjct: 203 SLTIDGIVYVIDPGFAKQKVYNPRVRVESLLVSPISKASAHQRSGRAGRTQPGKCFRLYT 382
Query: 249 SAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPF 289
+F+N+ + E+ + + VL LK + I + +F F
Sbjct: 383 EKSFNNDLQPQTYPEILRSNLANTVLTLKKLGIDDLVHFDF 505
>BM188234
Length = 424
Score = 96.7 bits (239), Expect = 4e-20
Identities = 52/138 (37%), Positives = 83/138 (59%)
Frame = +3
Query: 181 VATNVAETSLTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAA 240
+ATN+AETS+TIPGIKYV+D G K ++YD GME+ + SK+ A QR+GRAGR
Sbjct: 3 LATNIAETSVTIPGIKYVIDPGFVKARSYDPGKGMESLIIIPASKSQALQRSGRAGREGP 182
Query: 241 GHCYRLYSSAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLE 300
G C+RLY F + + + E+++ + V+L LK++ + + F F A++++
Sbjct: 183 GKCFRLYPEREF-EKLEDSTMPEIKRCNLSNVILQLKALGVDDILGFDFIEKPSRAAIIK 359
Query: 301 AENCLRALEALDSKDELT 318
+ L L AL + +L+
Sbjct: 360 SLEQLFLLGALTDECQLS 413
>BG881549 homologue to PIR|T48023|T48 ATP-dependent RNA helicase-like protein
- Arabidopsis thaliana, partial (15%)
Length = 434
Score = 80.1 bits (196), Expect = 3e-15
Identities = 48/100 (48%), Positives = 64/100 (64%), Gaps = 7/100 (7%)
Frame = +2
Query: 156 LYAMLPAAAQLRVFDG----VKEGE---RLVVVATNVAETSLTIPGIKYVVDTGREKVKN 208
+Y+ LP A Q ++F VKEG R ++V+T +AETSLTI I YV+D G K +
Sbjct: 32 VYSTLPPAMQQKIFKPAPPPVKEGGHPGRKIMVSTKIAETSLTIDSIVYVIDPGFAKQQV 211
Query: 209 YDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYS 248
Y+ +E+ V ISKASA QR+GRAGRT G C+RLY+
Sbjct: 212 YNPRIRVESLLVSPISKASAHQRSGRAGRTQPGKCFRLYT 331
>BG881871
Length = 431
Score = 73.9 bits (180), Expect = 2e-13
Identities = 41/106 (38%), Positives = 61/106 (56%)
Frame = +2
Query: 233 GRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTS 292
GR GRT G C++LY++ F E + + E+++ + VVL LK + I V +F F
Sbjct: 2 GRCGRTGPGKCFQLYTAYTFHKEMDDNTVPEIQRTNLANVVLTLKCLGIDNVMHFDFMDP 181
Query: 293 LKAASLLEAENCLRALEALDSKDELTLLGKAMALYPLSPRHSRMIL 338
+LL+A L AL AL+ ELT +G+ MA +PL P S+MI+
Sbjct: 182 PSDDALLKALELLYALSALNKFGELTKVGRRMAEFPLDPTLSKMIV 319
>TC210026 homologue to UP|Q9LXT8 (Q9LXT8) Pre-mRNA splicing factor
ATP-dependent RNA helicase-like protein, partial (20%)
Length = 750
Score = 67.4 bits (163), Expect = 2e-11
Identities = 56/231 (24%), Positives = 99/231 (42%)
Frame = +2
Query: 243 CYRLYSSAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAE 302
CYR+Y+ +A+ NE E+++ + VVLLLKS++++ + +F F ++L +
Sbjct: 2 CYRVYTESAYLNEMLPSPVPEIQRTNLGNVVLLLKSLKVENLLDFDFMDPPPQDNILNSM 181
Query: 303 NCLRALEALDSKDELTLLGKAMALYPLSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAA 362
L L AL++ LT LG M +PL P ++M+L + L +
Sbjct: 182 YQLWVLGALNNVGGLTDLGWKMVEFPLDPPLAKMLL----------MGEQLGCLEEVLTI 331
Query: 363 AAALSLPNPFVMQYEGNDSNKDSETSEKSRMGDNENNIDKTEKTKRKKLKQTSKVAREKF 422
+ LS+P+ F + K + S ARE+F
Sbjct: 332 VSMLSVPSVFF---------------------------------RPKDRAEESDAARERF 412
Query: 423 RIVSSDALAIAYALQCFEHSQNSVQFCEDNALHFKTMDEMSKLRQQLLRLV 473
+ SD L + Q ++ +C D+ LH K + + ++R QLL ++
Sbjct: 413 FVPESDHLTLYNVYQQWKQHDYRGDWCNDHFLHVKGLRKAREVRSQLLDIL 565
>TC219940 similar to UP|O49516 (O49516) RNA helicase - like protein, partial
(10%)
Length = 854
Score = 50.4 bits (119), Expect = 3e-06
Identities = 45/204 (22%), Positives = 87/204 (42%)
Frame = +2
Query: 411 LKQTSKVAREKFRIVSSDALAIAYALQCFEHSQNSVQFCEDNALHFKTMDEMSKLRQQLL 470
+++ S A+ +F D + + F S S Q+C N +++ M ++ ++R+QL
Sbjct: 53 IQKESDEAKLRFAAAEGDHVTFLNVYKGFHQSGKSSQWCHKNYVNYHAMRKVLEVREQLK 232
Query: 471 RLVFFQNDKGGLEQEYSWTHVTLEDVEHVWRVSSAHYPLPLVEERLICRAICAGWADRVA 530
R+ + GL L+ E + +++ +A+ AG+
Sbjct: 233 RIA----KRIGL---------VLKSCES--------------DMQVVRKAVTAGF----- 316
Query: 531 KRIPISKAVDGETISRAGRYQSCMVDESIFIHRWSSVSTVHPEFLVYNELLETKRPNKEG 590
+ A E S G Y++ + ++IH S + V+P++++YN L+ T
Sbjct: 317 ----FANACHLEAYSHNGMYKTLRGSQEVYIHPSSVLFRVNPKWVIYNSLVST------- 463
Query: 591 ETSAKRAYMHGVTNVDPTWLVENA 614
R YM V +DP+ L+E A
Sbjct: 464 ----DRQYMRNVITIDPSCLLEAA 523
>TC234889 weakly similar to UP|Q9SZ53 (Q9SZ53) Protein phosphatase 2C-like
protein (AT4g31860/F11C18_60), partial (10%)
Length = 661
Score = 45.8 bits (107), Expect(2) = 2e-05
Identities = 24/36 (66%), Positives = 26/36 (71%)
Frame = +1
Query: 277 KSMQIKKVANFPFPTSLKAASLLEAENCLRALEALD 312
KS VANF FPTSLK SLLE E CL+ALEAL+
Sbjct: 379 KSSMHLSVANFLFPTSLKDYSLLEVETCLKALEALE 486
Score = 21.2 bits (43), Expect(2) = 2e-05
Identities = 8/16 (50%), Positives = 13/16 (81%)
Frame = +3
Query: 312 DSKDELTLLGKAMALY 327
D+KDEL+ GKA++ +
Sbjct: 486 DNKDELSCSGKAISKF 533
>CD400081
Length = 635
Score = 44.7 bits (104), Expect(2) = 4e-05
Identities = 22/29 (75%), Positives = 24/29 (81%)
Frame = -1
Query: 284 VANFPFPTSLKAASLLEAENCLRALEALD 312
VANF FPTSLK SLLE E CL+ALEAL+
Sbjct: 407 VANFLFPTSLKDYSLLEVETCLKALEALE 321
Score = 21.2 bits (43), Expect(2) = 4e-05
Identities = 8/16 (50%), Positives = 13/16 (81%)
Frame = -3
Query: 312 DSKDELTLLGKAMALY 327
D+KDEL+ GKA++ +
Sbjct: 321 DNKDELSCSGKAISKF 274
>TC218921 weakly similar to GB|AAP37732.1|30725420|BT008373 At1g14690
{Arabidopsis thaliana;} , partial (9%)
Length = 276
Score = 44.3 bits (103), Expect = 2e-04
Identities = 18/24 (75%), Positives = 22/24 (91%)
Frame = +1
Query: 603 TNVDPTWLVENAKSSCIFSPPLTD 626
T+V+P WLVE+AKSSCIFSPPL +
Sbjct: 1 TSVEPAWLVEHAKSSCIFSPPLME 72
>TC229699 similar to UP|Q93Y39 (Q93Y39) ATP-dependent RNA helicase
(Fragment), partial (28%)
Length = 1742
Score = 39.7 bits (91), Expect = 0.005
Identities = 27/88 (30%), Positives = 41/88 (45%)
Frame = +2
Query: 153 VLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDSS 212
V L+A + A+L+ D +E E ++VAT+VA L IPG++ VV +Y
Sbjct: 356 VWTLHAQMQQRARLKAMDRFRENENGILVATDVAARGLDIPGVRTVV--------HYQLP 511
Query: 213 NGMETYEVKWISKASAAQRAGRAGRTAA 240
+ E Y R+GR R +A
Sbjct: 512 HSAEVY----------VHRSGRTARASA 565
>TC212408 similar to UP|Q867T7 (Q867T7) P67-like superoxide-generating NADPH
oxidase, partial (5%)
Length = 604
Score = 37.7 bits (86), Expect = 0.020
Identities = 23/73 (31%), Positives = 40/73 (54%)
Frame = -3
Query: 28 EINEAFEMPGSSSMQQTDRFSGYDEDDNNFDENESDSYDSETESELEFNDDDKNNHEGSK 87
E E +E GS + ++ + G DE D D E +++D E E+E D+D ++ + ++
Sbjct: 563 EDTEGYEDLGSDAQKR--KLQGTDEVDYE-DGPEEETHDGELSEEIE-GDEDGSDVDANE 396
Query: 88 NNNNIVDVLGNEG 100
N NN+ D +EG
Sbjct: 395 NYNNVTDANNSEG 357
>TC210714 weakly similar to UP|Q9VIZ3 (Q9VIZ3) CG10689-PA (LD25692p), partial
(9%)
Length = 599
Score = 36.2 bits (82), Expect = 0.058
Identities = 23/72 (31%), Positives = 36/72 (49%), Gaps = 1/72 (1%)
Frame = -1
Query: 544 ISRAGRYQSCMVDESIFIHRWSSVSTVHPEFLVYNEL-LETKRPNKEGETSAKRAYMHGV 602
+ + G Y++ +++ IH ++ V P ++VY+EL L TK YM V
Sbjct: 593 LQKNGSYRTVKHSQTVHIHPSLGLAQVLPRWVVYHELVLSTKE------------YMRQV 450
Query: 603 TNVDPTWLVENA 614
T + P WLVE A
Sbjct: 449 TELKPEWLVEIA 414
>TC226496 homologue to UP|Q93VJ8 (Q93VJ8) AT5g11200/F2I11_90
(AT5g11170/F2I11_60), partial (98%)
Length = 1674
Score = 34.3 bits (77), Expect = 0.22
Identities = 21/82 (25%), Positives = 39/82 (46%)
Frame = +1
Query: 156 LYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDSSNGM 215
+++ + +L+ + G KEG++ ++VAT++ + I + V+ NYD +
Sbjct: 1000 IHSAMSQEERLKRYKGFKEGKQRILVATDLVGRGIDIERVNIVI--------NYDMPDSA 1155
Query: 216 ETYEVKWISKASAAQRAGRAGR 237
+TY R GRAGR
Sbjct: 1156 DTY----------LHRVGRAGR 1191
>TC226495 homologue to UP|Q93VJ8 (Q93VJ8) AT5g11200/F2I11_90
(AT5g11170/F2I11_60), partial (32%)
Length = 846
Score = 33.9 bits (76), Expect = 0.29
Identities = 21/82 (25%), Positives = 38/82 (45%)
Frame = +3
Query: 156 LYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDSSNGM 215
++ + +L+ + G KEG++ ++VAT++ + I + V+ NYD +
Sbjct: 90 IHXAMSQEERLKRYKGFKEGKQRILVATDLVGRGIDIERVNIVI--------NYDMPDSA 245
Query: 216 ETYEVKWISKASAAQRAGRAGR 237
+TY R GRAGR
Sbjct: 246 DTY----------LHRVGRAGR 281
>TC231230 weakly similar to GB|CAA65563.1|1403551|SCLACHXVI P2570 protein
{Saccharomyces cerevisiae;} , partial (3%)
Length = 842
Score = 33.5 bits (75), Expect = 0.37
Identities = 13/32 (40%), Positives = 22/32 (68%)
Frame = +2
Query: 51 DEDDNNFDENESDSYDSETESELEFNDDDKNN 82
DED++N ++ E DS D + EFNDD++++
Sbjct: 569 DEDNDNEEDEEEDSGDDDGSQSEEFNDDEEDS 664
>TC227639 similar to UP|E74B_DROME (P11536) Ecdysone-induced protein 74EF
isoform B (ETS-related protein E74B), partial (3%)
Length = 1387
Score = 33.1 bits (74), Expect = 0.49
Identities = 14/43 (32%), Positives = 23/43 (52%)
Frame = +1
Query: 49 GYDEDDNNFDENESDSYDSETESELEFNDDDKNNHEGSKNNNN 91
G +ED++N DE D Y+SE + E + + KN + + N
Sbjct: 979 GVEEDEDNEDEEYDDDYESEYSEDEEDDQNTKNKQQAANGTVN 1107
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.130 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,055,944
Number of Sequences: 63676
Number of extensions: 463371
Number of successful extensions: 2712
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 2546
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2646
length of query: 789
length of database: 12,639,632
effective HSP length: 105
effective length of query: 684
effective length of database: 5,953,652
effective search space: 4072297968
effective search space used: 4072297968
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC147012.5


