

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.6 + phase: 0
(663 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
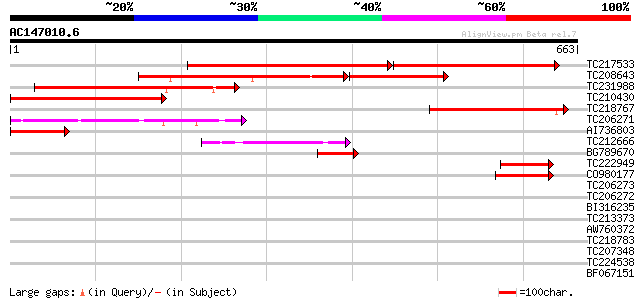
Score E
Sequences producing significant alignments: (bits) Value
TC217533 similar to UP|Q9LUS2 (Q9LUS2) Chloroplast outer envelop... 405 0.0
TC208643 similar to UP|Q41010 (Q41010) GTP-binding protein, part... 236 2e-93
TC231988 weakly similar to UP|Q7DLK2 (Q7DLK2) Chloroplast outer ... 223 2e-58
TC210430 weakly similar to UP|Q6RJN8 (Q6RJN8) Chloroplast Toc125... 186 4e-47
TC218767 similar to UP|Q41010 (Q41010) GTP-binding protein, part... 147 2e-35
TC206271 similar to UP|TO34_PEA (Q41009) Translocase of chloropl... 133 2e-31
AI736803 131 8e-31
TC212666 weakly similar to UP|Q6S5G3 (Q6S5G3) Chloroplast import... 107 1e-23
BG789670 60 2e-09
TC222949 47 4e-05
CO980177 46 6e-05
TC206273 similar to UP|TO34_PEA (Q41009) Translocase of chloropl... 40 0.003
TC206272 homologue to UP|TO34_PEA (Q41009) Translocase of chloro... 39 0.010
BI316235 similar to GP|10092443|gb| disease resistance protein A... 37 0.028
TC213373 similar to UP|Q9FIL7 (Q9FIL7) Similarity to protein kin... 34 0.18
AW760372 33 0.41
TC218783 similar to UP|Q40363 (Q40363) NuM1 protein, partial (19%) 32 0.69
TC207348 homologue to UP|Q40207 (Q40207) RAB1Y (Fragment), parti... 32 0.69
TC224538 32 0.91
BF067151 weakly similar to PIR|T06446|T064 GTP-binding protein -... 31 1.5
>TC217533 similar to UP|Q9LUS2 (Q9LUS2) Chloroplast outer envelope
protein-like (Chloroplast outer envelope
membrane-associated protein Toc120), partial (35%)
Length = 1633
Score = 405 bits (1040), Expect(2) = 0.0
Identities = 200/241 (82%), Positives = 218/241 (89%), Gaps = 1/241 (0%)
Frame = +1
Query: 208 NGQVWKPQLLLLSFASKILAEANALLKLQDNPREKPYTARARAPPLPFLLSSLLQSRPQL 267
NGQVWKP LLLLSFASKILAEANALLKLQD+P KPY AR RAPPLPFLLS+LLQSRPQL
Sbjct: 1 NGQVWKPHLLLLSFASKILAEANALLKLQDSPPGKPYVARTRAPPLPFLLSTLLQSRPQL 180
Query: 268 KLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQIRNLSRAQKKAYLDEVEY 327
KLPE+QF DEDSL+DDLDE S+S DE + DDLPPFKPLTKAQ+ LS+A KKAY DE+EY
Sbjct: 181 KLPEEQFGDEDSLDDDLDESSESDDENEHDDLPPFKPLTKAQVEKLSKAHKKAYFDELEY 360
Query: 328 REKLFMKKQLKYEKKQRKMMKEMAESVKDLPSDYVENVEEESGGAASVPVPMPDMSLPAS 387
REKL MKKQLK EKK+RKM+K+MAES KDLPSD+ ENVEEESGGAASVPVPMPD++LPAS
Sbjct: 361 REKLLMKKQLKEEKKRRKMLKKMAESAKDLPSDHSENVEEESGGAASVPVPMPDLALPAS 540
Query: 388 FDSDTPTHRYRHLD-SSNQWLVRPVLETHGWDHDVGYEGLNVERLFVLKDKIPVSFSGQV 446
FDSD PTHRYR+LD SSNQWLVRPVLETHGWDHDVGYEGLNVERLF LK+ IP+S SGQV
Sbjct: 541 FDSDNPTHRYRYLDSSSNQWLVRPVLETHGWDHDVGYEGLNVERLFDLKENIPLSVSGQV 720
Query: 447 T 447
T
Sbjct: 721 T 723
Score = 286 bits (733), Expect(2) = 0.0
Identities = 149/194 (76%), Positives = 167/194 (85%)
Frame = +2
Query: 449 DKKDANVQMEMTSSVKYGEGKATSLGFDMQTVGKDLAYTLRSETKFCNFLRNKATAGLSF 508
DK DA+V + S G+ ATS+ D QTV KDLAYTL ET F N N ATAGLSF
Sbjct: 728 DK*DADVTRAIYVSDMNGKWIATSICCDWQTVEKDLAYTLHCET*FTNMPLNNATAGLSF 907
Query: 509 TLLGDALSAGVKVEDKLIANKRFKLVIAGGAMTGRDDVAYGGSLEAQLRDKNYPLGRSLS 568
TLLGDALS+G+K+EDKL+A+KRFKLV++GGAMTGR D+AYGGSLEAQLRDK+YPLGR L+
Sbjct: 908 TLLGDALSSGLKIEDKLVASKRFKLVVSGGAMTGRGDIAYGGSLEAQLRDKDYPLGRFLT 1087
Query: 569 TLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALI 628
TLGLSVMDWHGDLAVG N+QSQIP+GRYTNLVARANLNNRGAGQISIRLNSSEQLQIALI
Sbjct: 1088TLGLSVMDWHGDLAVGYNVQSQIPVGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALI 1267
Query: 629 GLIPLLKKVIGYSQ 642
GLIPLLKK++GY Q
Sbjct: 1268GLIPLLKKLVGYHQ 1309
>TC208643 similar to UP|Q41010 (Q41010) GTP-binding protein, partial (42%)
Length = 1122
Score = 236 bits (602), Expect(2) = 2e-93
Identities = 125/256 (48%), Positives = 170/256 (65%), Gaps = 10/256 (3%)
Frame = +1
Query: 151 GPNGTPSSYDMFVTQRSHVVQQAIRQAAGDMRLMNP-----VSLVENHSACRTNTAGQRV 205
G G P SY++FV QRSH VQQ I QA GD+RLMNP VSLVENH +CR N GQ+V
Sbjct: 1 GTRGAPLSYEVFVAQRSHTVQQTIGQAVGDLRLMNPSLMNPVSLVENHPSCRKNRDGQKV 180
Query: 206 LPNGQVWKPQLLLLSFASKILAEANALLKLQDN-PREKPYTARARAPPLPFLLSSLLQSR 264
LPNGQ W+P LLLL ++ KIL+EA+ K Q++ + + R R+PPLP+LLSSLLQ+
Sbjct: 181 LPNGQSWRPLLLLLCYSMKILSEASNSTKTQESFDHRRLFGFRPRSPPLPYLLSSLLQTH 360
Query: 265 PQLKLPEDQFSDEDSLND----DLDEPSDSGDETDPDDLPPFKPLTKAQIRNLSRAQKKA 320
KLP DQ ++ +D DL + DE + D LPPFKP+ K+Q+ L++ Q+KA
Sbjct: 361 TYPKLPADQSGPDNGDSDVEMADLSDSDLDEDEDEYDQLPPFKPMKKSQVAKLTKEQQKA 540
Query: 321 YLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSDYVENVEEESGGAASVPVPMP 380
Y DE +YR KL KKQ + E ++ + MK+ + K+ Y+E ++E+G A+VPVP+P
Sbjct: 541 YFDEYDYRVKLLQKKQWREELRRMREMKKKG-NTKENDYGYMEEDDQENGSPAAVPVPLP 717
Query: 381 DMSLPASFDSDTPTHR 396
DM++P SFDSD P +R
Sbjct: 718 DMAMPPSFDSDNPAYR 765
Score = 125 bits (314), Expect(2) = 2e-93
Identities = 56/116 (48%), Positives = 82/116 (70%)
Frame = +2
Query: 398 RHLDSSNQWLVRPVLETHGWDHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQM 457
R L+ ++Q L RPVL+ HGWDHD GY+G+N+E+ + +K P + + VTKDKKD +Q+
Sbjct: 770 RFLEPTSQLLTRPVLDNHGWDHDCGYDGVNIEQSLAIINKFPAAVTVHVTKDKKDFTIQL 949
Query: 458 EMTSSVKYGEGKATSLGFDMQTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGD 513
+ + + K GE + GFD+Q+VGK L+Y++R ETK NF RNK +AG+S T LG+
Sbjct: 950 DSSVAAKLGENGSAMAGFDIQSVGKQLSYSVRGETKLKNFKRNKTSAGVSVTYLGE 1117
>TC231988 weakly similar to UP|Q7DLK2 (Q7DLK2) Chloroplast outer envelope
protein 86, partial (22%)
Length = 950
Score = 223 bits (569), Expect = 2e-58
Identities = 121/247 (48%), Positives = 166/247 (66%), Gaps = 8/247 (3%)
Frame = +3
Query: 30 SSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDTPGLLPSWSDQPHNEKILH 89
S+TINSIF ++K T+AF T V++V G + GIK+R++DTPGL +Q +N KIL
Sbjct: 3 SATINSIFGDMKVVTNAFEPATTSVKEVSGTIDGIKIRILDTPGLKSCIKEQAYNRKILS 182
Query: 90 SVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIFGPPIWFNAIVVLTHAASAPP 149
SVKR++KK PPD++LY+DR+D Q+RD +D+P+LR+IT GP IW +AI+ LTHAAS P
Sbjct: 183 SVKRYMKKFPPDVILYVDRVDAQTRDLNDLPILRSITSSLGPSIWQHAILTLTHAASVPF 362
Query: 150 DGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDM-----RLMNPVSLVENHSACRTNTAGQR 204
DGP+G+P SY++FV Q+S++VQQ+I QA ++ M PV+LVENH C N G
Sbjct: 363 DGPSGSPLSYEVFVAQKSYLVQQSITQAVRNLCQLSPSFMCPVALVENHPLCGKNMFGDC 542
Query: 205 VLPNGQVWKPQLLLLSFASKILAEANALLKLQ---DNPREKPYTARARAPPLPFLLSSLL 261
VLPNG W+ QLL L F+ KI++E +++ Q DN K + PL L SSLL
Sbjct: 543 VLPNGLRWRSQLLALCFSLKIVSEVSSISGSQTLFDN--WKNCFFQDHPQPLCHLFSSLL 716
Query: 262 QSRPQLK 268
+S LK
Sbjct: 717 KSPAHLK 737
>TC210430 weakly similar to UP|Q6RJN8 (Q6RJN8) Chloroplast Toc125, partial
(9%)
Length = 567
Score = 186 bits (471), Expect = 4e-47
Identities = 88/184 (47%), Positives = 131/184 (70%), Gaps = 1/184 (0%)
Frame = +2
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
+A + E+ G LDFSC I+VLGK+GVGKS+TINSIF + K T AF T +Q+VVG
Sbjct: 14 IASEQEATGMPQLDFSCRILVLGKTGVGKSATINSIFGQAKTTTGAFQPATNCIQEVVGN 193
Query: 61 VQGIKVRVIDTPGLLPS-WSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDM 119
V G+ + IDTPG LPS ++ N++I+ S+KRFI+K+PPDIVLY +RLD + + D
Sbjct: 194 VNGLNIAFIDTPGFLPSSTNNMKRNKRIMLSIKRFIRKSPPDIVLYFERLDFINAGYVDF 373
Query: 120 PLLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAG 179
PLL+ +T++FG IWFN I+V+TH++SA P+GP+G +Y+ +V+ ++++Q I+Q
Sbjct: 374 PLLKLVTEVFGSAIWFNTIIVMTHSSSAIPEGPDGYTFNYESYVSYCTNMIQLHIQQVVF 553
Query: 180 DMRL 183
D ++
Sbjct: 554 DSKV 565
>TC218767 similar to UP|Q41010 (Q41010) GTP-binding protein, partial (19%)
Length = 748
Score = 147 bits (370), Expect = 2e-35
Identities = 75/168 (44%), Positives = 113/168 (66%), Gaps = 5/168 (2%)
Frame = +1
Query: 491 ETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAMTGRDDVAYGG 550
ETK NF RNK +AG+S T LG+ + G+KVED++ KR LV + G + + D AYG
Sbjct: 4 ETKLKNFKRNKTSAGVSVTYLGENVCTGLKVEDQIAVGKRLVLVGSTGVVKSKTDSAYGA 183
Query: 551 SLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLVARANLNNRGA 610
++E +LR+ ++P+G+ S+L LS++ W GDLA+G NLQSQ +GR + RA LNN+ +
Sbjct: 184 NVEVRLREADFPIGQDQSSLSLSLVKWRGDLALGANLQSQFSVGRGYKVAVRAGLNNKLS 363
Query: 611 GQISIRLNSSEQLQIALIGLIPLLKKVI-----GYSQNCSLDKIIDEV 653
GQI++R +SS+QLQIAL+ ++P+ K + G S+N S+ I+ V
Sbjct: 364 GQITVRTSSSDQLQIALVAILPIAKAIYKNFWPGASENYSIY*IVSFV 507
>TC206271 similar to UP|TO34_PEA (Q41009) Translocase of chloroplast 34 (34
kDa chloroplast outer envelope protein) (GTP-binding
protein OEP34) (GTP-binding protein IAP34), partial
(97%)
Length = 1345
Score = 133 bits (335), Expect = 2e-31
Identities = 98/288 (34%), Positives = 138/288 (47%), Gaps = 11/288 (3%)
Frame = +2
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
+ E L QE ++ S TI+V+GK GVGKSST+NSI + + + F + V
Sbjct: 224 LLELLGKLKQENVN-SLTILVMGKGGVGKSSTVNSIIGDRVVSINPFQSEGPRPVIVSRS 400
Query: 61 VQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP 120
G + +IDTPGL+ N+ L +KRF+ D++LY+DRLD+ D D
Sbjct: 401 RAGFTLNIIDTPGLIEGGYI---NDMALDIIKRFLLNKTIDVLLYVDRLDVYRVDNLDKV 571
Query: 121 LLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAA-- 178
+ + ITD FG IW I+ LTHA +PPDG YD F +QRS + + +R A
Sbjct: 572 VAKAITDSFGKGIWSKTILALTHAQFSPPDG-----LPYDEFFSQRSESLLKVLRSGARI 736
Query: 179 ---GDMRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQL------LLLSFASKILAEA 229
PV LVEN C N + ++VLPNG W P L + L+ + I +
Sbjct: 737 KKEAFQAASIPVVLVENSGRCNKNDSDEKVLPNGTAWIPNLVQTITEIALNKSESIHVDK 916
Query: 230 NALLKLQDNPREKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDE 277
N + N R K + PL F L L +P L E ++E
Sbjct: 917 NLIEGPNPNQRGKLWI------PLVFALQYFLIMKPIKGLIEKDIANE 1042
>AI736803
Length = 411
Score = 131 bits (330), Expect = 8e-31
Identities = 65/70 (92%), Positives = 68/70 (96%)
Frame = +3
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
MAEQLE+AGQEPLDFSCTIMVLGK+GVGKS+TINSIFDEVKFNT AFHMGTKKVQDVVG
Sbjct: 201 MAEQLEAAGQEPLDFSCTIMVLGKTGVGKSATINSIFDEVKFNTSAFHMGTKKVQDVVGT 380
Query: 61 VQGIKVRVID 70
VQGIKVRVID
Sbjct: 381 VQGIKVRVID 410
>TC212666 weakly similar to UP|Q6S5G3 (Q6S5G3) Chloroplast import receptor
Toc90, partial (14%)
Length = 490
Score = 107 bits (268), Expect = 1e-23
Identities = 70/176 (39%), Positives = 102/176 (57%), Gaps = 2/176 (1%)
Frame = +3
Query: 225 ILAEANALLKLQDNPREKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDL 284
+L + N+LLK Q++ P + R P +P LLSSLL+ R S+ +D++
Sbjct: 3 VLGDVNSLLKFQNSVELGPLNS-PRIPSMPHLLSSLLRHR--------LVSNLSGTDDEI 155
Query: 285 DEP--SDSGDETDPDDLPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKK 342
+E SD +E + D LP + LTK+Q L KK YLDE++YRE L++KKQLK E
Sbjct: 156 EEILLSDKKEEDEYDQLPSIRVLTKSQFEKLPEPLKKDYLDEMDYRETLYLKKQLK-EDY 332
Query: 343 QRKMMKEMAESVKDLPSDYVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYR 398
QR+ K ++ K L D ++ + A + PV +PDM++PASFDSD +HRYR
Sbjct: 333 QRRKEKLLSTDKKFLNGDNPDDQQ-----APTEPVLLPDMAVPASFDSDCHSHRYR 485
>BG789670
Length = 348
Score = 60.5 bits (145), Expect = 2e-09
Identities = 25/47 (53%), Positives = 37/47 (78%)
Frame = +1
Query: 361 YVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWL 407
Y+E ++E+G A+VPVP+PDM++P SFDSD P +RYR L+ ++Q L
Sbjct: 208 YMEEDDQENGSPAAVPVPLPDMAMPPSFDSDNPAYRYRFLNPTSQLL 348
Score = 32.7 bits (73), Expect(2) = 0.048
Identities = 12/23 (52%), Positives = 18/23 (78%)
Frame = +2
Query: 297 DDLPPFKPLTKAQIRNLSRAQKK 319
D LPPFKP+ K+Q+ L++ Q+K
Sbjct: 128 DQLPPFKPMKKSQVAKLTKEQQK 196
Score = 22.3 bits (46), Expect(2) = 0.048
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 1/34 (2%)
Frame = +1
Query: 258 SSLLQSRPQLKLPEDQFSDEDSLND-DLDEPSDS 290
+SLLQ+ KLP DQ ++ +D ++ + SDS
Sbjct: 1 ASLLQTHTYPKLPADQSGPDNGDSDVEMADLSDS 102
>TC222949
Length = 608
Score = 46.6 bits (109), Expect = 4e-05
Identities = 22/62 (35%), Positives = 40/62 (64%)
Frame = +2
Query: 574 VMDWHGDLAVGCNLQSQIPIGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPL 633
V+ ++ ++ + +LQS+ + R + ANLN+R GQI I+++SSE LQIA + ++ +
Sbjct: 2 VLSFNKEMVLSGSLQSEFRLSRSSKASVSANLNSRKMGQICIKISSSEHLQIASVAILSI 181
Query: 634 LK 635
K
Sbjct: 182 WK 187
>CO980177
Length = 813
Score = 45.8 bits (107), Expect = 6e-05
Identities = 23/67 (34%), Positives = 42/67 (62%)
Frame = -1
Query: 569 TLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALI 628
+L ++V ++ ++ + +LQS+ + R + ANL +R GQI I+++SSE LQIA +
Sbjct: 795 SLTMTVXXFNKEMVLSGSLQSEFRLSRSSKASVSANLXSRKMGQICIKISSSEHLQIASV 616
Query: 629 GLIPLLK 635
+ +LK
Sbjct: 615 AVFSILK 595
>TC206273 similar to UP|TO34_PEA (Q41009) Translocase of chloroplast 34 (34
kDa chloroplast outer envelope protein) (GTP-binding
protein OEP34) (GTP-binding protein IAP34), partial
(36%)
Length = 732
Score = 40.0 bits (92), Expect = 0.003
Identities = 29/98 (29%), Positives = 42/98 (42%), Gaps = 6/98 (6%)
Frame = +1
Query: 186 PVSLVENHSACRTNTAGQRVLPNGQVWKPQL------LLLSFASKILAEANALLKLQDNP 239
PV LVEN C N + ++ LPNG W P L + L+ + I + + N
Sbjct: 34 PVVLVENSGRCNKNDSDEKXLPNGTAWIPNLVQTITDIALNQSESIHVDKXXIEGPNPNQ 213
Query: 240 REKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDE 277
R K +T PL F + L +P L E ++E
Sbjct: 214 RGKLWT------PLVFAIQYFLIMKPIKGLIEKDIANE 309
>TC206272 homologue to UP|TO34_PEA (Q41009) Translocase of chloroplast 34 (34
kDa chloroplast outer envelope protein) (GTP-binding
protein OEP34) (GTP-binding protein IAP34), partial
(19%)
Length = 375
Score = 38.5 bits (88), Expect = 0.010
Identities = 21/39 (53%), Positives = 28/39 (70%)
Frame = +3
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDE 39
+ E L + QE ++ S TI+V+GK GVGKSST+NSI E
Sbjct: 243 LLELLGNLKQENVN-SLTILVMGKGGVGKSSTVNSIIGE 356
>BI316235 similar to GP|10092443|gb| disease resistance protein AIG1;
5333-4002 {Arabidopsis thaliana}, partial (25%)
Length = 409
Score = 37.0 bits (84), Expect = 0.028
Identities = 22/59 (37%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Frame = +1
Query: 18 TIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDV--VGMVQGIKVRVIDTPGL 74
T++++G++G GKS+T NS+ F + G V ++ M G V VIDTPGL
Sbjct: 160 TLVLVGRTGNGKSATGNSVLGRRAFKSRTSSSGVTSVCELQRTIMKDGSIVNVIDTPGL 336
>TC213373 similar to UP|Q9FIL7 (Q9FIL7) Similarity to protein kinase, partial
(17%)
Length = 832
Score = 34.3 bits (77), Expect = 0.18
Identities = 30/92 (32%), Positives = 44/92 (47%)
Frame = -2
Query: 252 PLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQIR 311
P+P LL L +P+D ED+L D+L E SD D + DD P L +
Sbjct: 435 PIPSLL---------LAVPDDAEE*EDTLFDELLELSD--DTSVEDDTP--LTLCVFFLS 295
Query: 312 NLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQ 343
+LS A + +L K F+K+ LKY + +
Sbjct: 294 SLSAASESVFLFPERNNVKNFLKEDLKYVRAE 199
>AW760372
Length = 423
Score = 33.1 bits (74), Expect = 0.41
Identities = 15/27 (55%), Positives = 21/27 (77%)
Frame = +3
Query: 2 AEQLESAGQEPLDFSCTIMVLGKSGVG 28
A++LE Q+ LDFS I++LG+SGVG
Sbjct: 342 AKKLEENCQDDLDFSLNILLLGRSGVG 422
>TC218783 similar to UP|Q40363 (Q40363) NuM1 protein, partial (19%)
Length = 870
Score = 32.3 bits (72), Expect = 0.69
Identities = 31/114 (27%), Positives = 50/114 (43%), Gaps = 1/114 (0%)
Frame = +3
Query: 258 SSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQIRNLSRAQ 317
S+ Q + + +D SDED D+D+ DS DE++ +P K ++N +
Sbjct: 519 SARAQKKVESSDSDDSSSDEDKGKMDIDDDDDSSDESE-------EPQKKKAVKNSKESS 677
Query: 318 KKAYLDEVEYREKLFMKKQLKYEKKQRKM-MKEMAESVKDLPSDYVENVEEESG 370
+ D + E+ K +K+ R + M + A S K P V EESG
Sbjct: 678 DSSEEDSEDESEE---KPSKTPQKRGRDVEMVDAALSEKKAPKTPV-TPREESG 827
>TC207348 homologue to UP|Q40207 (Q40207) RAB1Y (Fragment), partial (50%)
Length = 479
Score = 32.3 bits (72), Expect = 0.69
Identities = 23/73 (31%), Positives = 40/73 (54%), Gaps = 6/73 (8%)
Frame = +3
Query: 7 SAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFH-----MGTK-KVQDVVGM 60
S+G + D+ ++++G SGVGKSS + + F +DAF +G KV+ V+
Sbjct: 171 SSGHQEFDYLFKLLMIGDSGVGKSSLL------LSFTSDAFEDLSPTIGVDFKVKYVMMG 332
Query: 61 VQGIKVRVIDTPG 73
+ +K+ + DT G
Sbjct: 333 GKKLKLAIWDTAG 371
>TC224538
Length = 736
Score = 32.0 bits (71), Expect = 0.91
Identities = 20/68 (29%), Positives = 35/68 (51%)
Frame = +1
Query: 522 EDKLIANKRFKLVIAGGAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDL 581
EDK+ +NK +V ++ DD S E +L+ Y + S ++ S++D +GD+
Sbjct: 454 EDKIASNK---IVPISLPISESDDDESVMSSEGRLQVGRYQIKESFGSILQSILDKYGDI 624
Query: 582 AVGCNLQS 589
C L+S
Sbjct: 625 GESCQLES 648
>BF067151 weakly similar to PIR|T06446|T064 GTP-binding protein - garden pea,
partial (21%)
Length = 332
Score = 31.2 bits (69), Expect = 1.5
Identities = 17/49 (34%), Positives = 26/49 (52%)
Frame = +2
Query: 4 QLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTK 52
Q + E +D+ I++ G SGVGKS +N +F + FHM +K
Sbjct: 185 QWQGNADEGIDYMFKIVMTGDSGVGKSQLLN------RFVKNEFHMKSK 313
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,728,291
Number of Sequences: 63676
Number of extensions: 334743
Number of successful extensions: 2025
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 1956
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2005
length of query: 663
length of database: 12,639,632
effective HSP length: 103
effective length of query: 560
effective length of database: 6,081,004
effective search space: 3405362240
effective search space used: 3405362240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147010.6


