

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.5 - phase: 0
(657 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
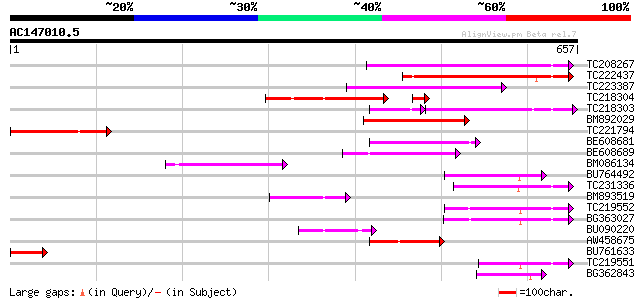
Score E
Sequences producing significant alignments: (bits) Value
TC208267 homologue to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, p... 155 4e-38
TC222437 weakly similar to UP|O23045 (O23045) YUP8H12.16 protein... 154 2e-37
TC223387 138 9e-33
TC218304 similar to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, par... 125 8e-31
TC218303 homologue to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, p... 112 1e-30
BM892029 116 4e-26
TC221794 homologue to UP|Q9FR59 (Q9FR59) Homeobox 1, partial (20%) 113 2e-25
BE608681 weakly similar to GP|3925363|gb| homeodomain protein {M... 88 1e-17
BE608689 similar to GP|3925363|gb| homeodomain protein {Malus x ... 88 1e-17
BM086134 85 9e-17
BU764492 79 8e-15
TC231336 similar to UP|Q9ZTA8 (Q9ZTA8) Homeodomain protein (Frag... 75 9e-14
BM893519 similar to GP|22475197|gb homeodomain protein GhHOX2 {G... 73 4e-13
TC219552 similar to UP|Q8LJS8 (Q8LJS8) Homeodomain protein GhHOX... 73 4e-13
BG363027 72 1e-12
BU090220 similar to GP|22475197|gb| homeodomain protein GhHOX2 {... 69 9e-12
AW458675 similar to GP|1881536|gb|A A20 {Arabidopsis thaliana}, ... 63 5e-10
BU761633 similar to GP|1814424|gb|A homeodomain protein AHDP {Ar... 57 3e-08
TC219551 similar to UP|Q8LJS8 (Q8LJS8) Homeodomain protein GhHOX... 53 5e-07
BG362843 similar to GP|8885530|dbj| homeodomain transcription fa... 51 2e-06
>TC208267 homologue to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, partial (31%)
Length = 1049
Score = 155 bits (393), Expect = 4e-38
Identities = 91/242 (37%), Positives = 136/242 (55%), Gaps = 2/242 (0%)
Frame = +2
Query: 414 QTLEGRNSVIKLAQRMVKMFCESLTMPGQLELNHLTLASIGGIRVSFRSTTDDDTSQPNG 473
QT GR S++KLA+RMV FC ++ L+ +RV R + DD +P G
Sbjct: 35 QTKTGRKSMMKLAERMVISFCAGVSASTAHTWTTLSGTGADDVRVMTRKSADDP-GRPPG 211
Query: 474 TIVTAATTLWLPLPALKVFEFLKDPTKRSQWDGLSCGNPMHEIAHISNGPYHGNCISIIK 533
+++AAT+ WLP+P +VF+FL+D R++WD LS G + E+AHI+NG GNC+S+++
Sbjct: 212 IVLSAATSFWLPVPPKRVFDFLRDENSRNEWDILSNGGVVQEMAHIANGRDTGNCVSLLR 391
Query: 534 --PFIPTQRQMMILQESFTSRVGSYIIYAPSDRQTMDVALRGEDSKELPILPYGFVVCSK 591
+Q M+ILQES T+ GS++IYAP D M+V L G D + +LP GF +
Sbjct: 392 VNSANSSQSNMLILQESCTNSTGSFVIYAPVDIVAMNVVLNGGDPDYVALLPSGFAILPD 571
Query: 592 SQPNLNAPFGASNNIEDGSLLTLAAQILSTSPHEIDQVLNVEDITDINTHLATTILNVKD 651
+ N G GSLLT+A QIL S L++ + +N +A T+ +K
Sbjct: 572 GTTSHNGSGGIGETGPSGSLLTVAFQILVDSVPTAK--LSLGSVATVNNLIACTVERIKA 745
Query: 652 AL 653
+L
Sbjct: 746 SL 751
>TC222437 weakly similar to UP|O23045 (O23045) YUP8H12.16 protein, partial
(16%)
Length = 719
Score = 154 bits (388), Expect = 2e-37
Identities = 81/207 (39%), Positives = 125/207 (60%), Gaps = 9/207 (4%)
Frame = +1
Query: 456 IRVSFRSTTDDDTSQPNGTIVTAATTLWLPLPALKVFEFLKDPTKRSQWDGLSCGNPMHE 515
+RV+ ++D QPNG +++AATT+WLP P VF F KD KR QWD LS GN + E
Sbjct: 1 VRVTVHKSSDP--GQPNGVVLSAATTIWLPTPPHAVFNFFKDENKRPQWDVLSNGNAVQE 174
Query: 516 IAHISNGPYHGNCISIIKPFIPTQRQMMILQESFTSRVGSYIIYAPSDRQTMDVALRGED 575
+A+I+NG + GN IS+++ F + + M+ILQES GS+++Y P D ++++A+ GED
Sbjct: 175 VANIANGLHPGNSISVLRAFNNSTQNMLILQESCIDSYGSFVVYCPVDLPSINLAMSGED 354
Query: 576 SKELPILPYGFVVCSKSQPNLNAPFGASNNIED---------GSLLTLAAQILSTSPHEI 626
+P+LP GF + QP+ GAS + + GSL+T+A QIL +S
Sbjct: 355 PSYIPLLPNGFTILPDGQPDQEGDDGASTSSNNANRNIVRSGGSLVTIAFQILVSSLPSA 534
Query: 627 DQVLNVEDITDINTHLATTILNVKDAL 653
LN+E +T +N + +T+ +K +L
Sbjct: 535 K--LNMESVTTVNNLIGSTVQQIKSSL 609
>TC223387
Length = 569
Score = 138 bits (347), Expect = 9e-33
Identities = 76/187 (40%), Positives = 110/187 (58%), Gaps = 2/187 (1%)
Frame = +2
Query: 391 RMCERSLGSYVEAIPVEETIGVIQTLEGRNSVIKLAQRMVKMFCESLTMPGQLELNHLTL 450
R CER S IP + + VI + EGR S++KLA+RMV +C + L+
Sbjct: 2 RQCERLASSMANNIPAGD-LCVITSAEGRKSMMKLAERMVMSYCTGVGASTAHAWTTLSA 178
Query: 451 ASIGGIRVSFRSTTDDDTSQPNGTIVTAATTLWLPLPALKVFEFLKDPTKRSQWDGLSCG 510
+RV R +TD+ +P G +++AAT+ WLP+P +VF FL+D R++WD LS G
Sbjct: 179 TGCDDVRVMTRKSTDEP-GRPPGIVLSAATSFWLPVPPKRVFHFLRDQNSRNEWDILSNG 355
Query: 511 NPMHEIAHISNGPYHGNCISIIK--PFIPTQRQMMILQESFTSRVGSYIIYAPSDRQTMD 568
+ E+AHI+NG GNC+S+++ +Q M+ILQES T GSY++YAP D M+
Sbjct: 356 GLVQELAHIANGRDPGNCVSLLRVNSANSSQSNMLILQESCTDSTGSYVVYAPVDIVAMN 535
Query: 569 VALRGED 575
V L G D
Sbjct: 536 VVLSGGD 556
>TC218304 similar to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, partial (26%)
Length = 575
Score = 125 bits (315), Expect(2) = 8e-31
Identities = 63/142 (44%), Positives = 90/142 (63%)
Frame = +1
Query: 297 REFNIIRYCKKFDAGVWVIADVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVE 356
RE +RYCK+ G W + DVS D+ RP+ P +R + PSGC+I+EMP+G V WVE
Sbjct: 7 REIYFVRYCKQHGDGTWAVVDVSLDNLRPS--PSARCRRRPSGCLIQEMPNGYSKVIWVE 180
Query: 357 HVEVEDKIHTHYVYRDLVGNYNLYGAESWIKELQRMCERSLGSYVEAIPVEETIGVIQTL 416
HVEV+D+ H +Y+ LV + + +GA+ WI L R CER + IP + +GVI
Sbjct: 181 HVEVDDR-GVHNLYKQLVSSGHAFGAKRWIANLDRQCERLASAMATNIPTVD-VGVITNP 354
Query: 417 EGRNSVIKLAQRMVKMFCESLT 438
+GR S++KLA+RMV FC ++
Sbjct: 355 DGRKSMLKLAERMVISFCAGVS 420
Score = 26.9 bits (58), Expect(2) = 8e-31
Identities = 9/20 (45%), Positives = 15/20 (75%)
Frame = +2
Query: 467 DTSQPNGTIVTAATTLWLPL 486
D +P G +++AAT+ WLP+
Sbjct: 503 DPGRPPGIVLSAATSFWLPV 562
>TC218303 homologue to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, partial (32%)
Length = 1230
Score = 112 bits (281), Expect(2) = 1e-30
Identities = 67/177 (37%), Positives = 97/177 (53%), Gaps = 2/177 (1%)
Frame = +2
Query: 483 WLPLPALKVFEFLKDPTKRSQWDGLSCGNPMHEIAHISNGPYHGNCISIIK--PFIPTQR 540
WLP+ +VFEFL+D RS+WD LS G + E+AHI+NG GNC+S+++ +Q
Sbjct: 206 WLPVSPKRVFEFLRDENSRSEWDILSNGGVVQEMAHIANGRDTGNCVSLLRVNSANSSQS 385
Query: 541 QMMILQESFTSRVGSYIIYAPSDRQTMDVALRGEDSKELPILPYGFVVCSKSQPNLNAPF 600
M+ILQES GS++IYAP D M+V L G D + +LP GF +
Sbjct: 386 NMLILQESCADSTGSFVIYAPVDIVAMNVVLNGGDPDYVALLPSGFAILPDGTTAHGGGI 565
Query: 601 GASNNIEDGSLLTLAAQILSTSPHEIDQVLNVEDITDINTHLATTILNVKDALMSSI 657
G + + GSLLT+A QIL S L++ + +N +A T+ +K AL +
Sbjct: 566 GDTGH--GGSLLTVAFQILVDSVPTAK--LSLGSVATVNNLIACTVERIKAALSGEV 724
Score = 39.3 bits (90), Expect(2) = 1e-30
Identities = 22/65 (33%), Positives = 36/65 (54%)
Frame = +3
Query: 417 EGRNSVIKLAQRMVKMFCESLTMPGQLELNHLTLASIGGIRVSFRSTTDDDTSQPNGTIV 476
+GR S++KLA+RMV FC ++ L+ +RV R + DD +P G ++
Sbjct: 12 DGRKSMLKLAERMVISFCAGVSASTAHTWTTLSGTGADDVRVMTRKSV-DDPGRPPGIVL 188
Query: 477 TAATT 481
+AAT+
Sbjct: 189 SAATS 203
>BM892029
Length = 421
Score = 116 bits (290), Expect = 4e-26
Identities = 56/123 (45%), Positives = 78/123 (62%)
Frame = +2
Query: 411 GVIQTLEGRNSVIKLAQRMVKMFCESLTMPGQLELNHLTLASIGGIRVSFRSTTDDDTSQ 470
GVI + EG+ S++KLAQRMV FC S++ L+ + + + V D Q
Sbjct: 50 GVIPSPEGKRSMMKLAQRMVTNFCASISSSAGHRWTTLSGSGMNEVGVRVTVHKSSDPGQ 229
Query: 471 PNGTIVTAATTLWLPLPALKVFEFLKDPTKRSQWDGLSCGNPMHEIAHISNGPYHGNCIS 530
PNG +++AATT+WLP+P VF F KD KR QWD LS GN + E+AHI+NG + GNCIS
Sbjct: 230 PNGVVLSAATTIWLPIPPQTVFNFFKDEKKRPQWDVLSNGNAVQEVAHIANGSHPGNCIS 409
Query: 531 IIK 533
+++
Sbjct: 410 VLR 418
>TC221794 homologue to UP|Q9FR59 (Q9FR59) Homeobox 1, partial (20%)
Length = 501
Score = 113 bits (283), Expect = 2e-25
Identities = 53/117 (45%), Positives = 80/117 (68%)
Frame = +3
Query: 2 KECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ +R +L+ E+GLEP Q+KFWFQNKRT +K QHER N LR EN+K+R +N+
Sbjct: 129 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKTQHERHENTQLRTENEKLRADNM 308
Query: 62 KIKEVLKAKICLDCGGPPFPMKDHQNFVQDLKQENAQLKQECEKMSSLLASYMEKKI 118
+ +E L C +CGGP + + L+ ENA+L++E +++S++ A Y+ K +
Sbjct: 309 RFREALGNASCPNCGGPT-AIGEMSFDEHHLRLENARLREEIDRISAIAAKYVGKPV 476
>BE608681 weakly similar to GP|3925363|gb| homeodomain protein {Malus x
domestica}, partial (19%)
Length = 475
Score = 88.2 bits (217), Expect = 1e-17
Identities = 42/128 (32%), Positives = 75/128 (57%)
Frame = +2
Query: 418 GRNSVIKLAQRMVKMFCESLTMPGQLELNHLTLASIGGIRVSFRSTTDDDTSQPNGTIVT 477
G S++KLAQ M +C ++ L+ ++L ++++G DD +P G ++T
Sbjct: 80 GGKSMLKLAQLMTSNYCSGVSASSALKWDNLHISTLGDDMKVMTRKNVDDPGEPTGIVLT 259
Query: 478 AATTLWLPLPALKVFEFLKDPTKRSQWDGLSCGNPMHEIAHISNGPYHGNCISIIKPFIP 537
AAT++W+P+ +++ L D RS+WD L G PMH++ HI HGNC+SI++ +P
Sbjct: 260 AATSVWVPVTRMRLLNSLLDEHLRSEWDILFNGGPMHDMVHIVR*QGHGNCVSILR-LMP 436
Query: 538 TQRQMMIL 545
++ + +L
Sbjct: 437 LRQLIYLL 460
>BE608689 similar to GP|3925363|gb| homeodomain protein {Malus x domestica},
partial (18%)
Length = 413
Score = 87.8 bits (216), Expect = 1e-17
Identities = 47/137 (34%), Positives = 74/137 (53%)
Frame = +2
Query: 386 IKELQRMCERSLGSYVEAIPVEETIGVIQTLEGRNSVIKLAQRMVKMFCESLTMPGQLEL 445
I LQR CE +I ++ + Q GR S++KLAQRM FC + +
Sbjct: 2 IATLQRQCECLAILMSSSISSDDHTALSQA--GRRSMLKLAQRMTSNFCSGVCASSARKW 175
Query: 446 NHLTLASIGGIRVSFRSTTDDDTSQPNGTIVTAATTLWLPLPALKVFEFLKDPTKRSQWD 505
+ L + ++G DD +P G +++AAT++W+P+ ++F+FL+D RS+WD
Sbjct: 176 DSLHIGTLGDDMKVMTRKNVDDPGEPPGIVLSAATSVWVPVSRQRLFDFLRDERLRSEWD 355
Query: 506 GLSCGNPMHEIAHISNG 522
LS G PM E+ HI+ G
Sbjct: 356 ILSNGGPMQEMVHIAKG 406
>BM086134
Length = 428
Score = 85.1 bits (209), Expect = 9e-17
Identities = 45/144 (31%), Positives = 83/144 (57%), Gaps = 2/144 (1%)
Frame = +1
Query: 181 VRLVRVNEPFWVKSPNTQDGYT--FHRESYEQVFPKNNHFKGANVCEESSKYSGLVKISG 238
+ + ++ EP W+ T DG + + + Y + FP+ K + E+S+ + +V ++
Sbjct: 1 IGMAQMGEPLWL---TTLDGTSTMLNEDEYIRSFPRGIGPKPSGFKCEASRETAVVIMNH 171
Query: 239 IDLVGMFLDSVKWTNLFPTIVTKAETIKVFEIGSPGSRDGALLLMNEEMHILSPLVRPRE 298
++LV + +D +W+ +F IV++A T++V G G+ +GAL +M E+ + +PLV RE
Sbjct: 172 VNLVEILMDVNQWSTVFSGIVSRAMTLEVLSTGVAGNYNGALQVMTAELQLPTPLVPTRE 351
Query: 299 FNIIRYCKKFDAGVWVIADVSFDS 322
+RYCK+ G W + DVS D+
Sbjct: 352 SYFVRYCKQHADGTWAVVDVSLDN 423
>BU764492
Length = 428
Score = 78.6 bits (192), Expect = 8e-15
Identities = 50/129 (38%), Positives = 69/129 (52%), Gaps = 11/129 (8%)
Frame = +3
Query: 505 DGLSCGNPMHEIAHISNGPYHGNCISIIKPFI--PTQRQMMILQESFTSRVGSYIIYAPS 562
D LS G PM E+AHI+ G HGN +S+++ Q M+ILQE+ GS ++YAP
Sbjct: 3 DILSNGGPMQEMAHIAKGQDHGNAVSLLRASAINSNQSSMLILQETCIDAAGSLVVYAPV 182
Query: 563 DRQTMDVALRGEDSKELPILPYGFVVC-----SKSQPN--LNAPFGASNNIE--DGSLLT 613
D + V + G DS + +LP GF + S+ PN + G N + GSLLT
Sbjct: 183 DIPVLYVVMNGGDSAYVALLPSGFAIVPDGPGSRGPPNGPTSTTNGGDNGVTRVSGSLLT 362
Query: 614 LAAQILSTS 622
+A QIL S
Sbjct: 363 VAFQILVNS 389
>TC231336 similar to UP|Q9ZTA8 (Q9ZTA8) Homeodomain protein (Fragment),
partial (19%)
Length = 869
Score = 75.1 bits (183), Expect = 9e-14
Identities = 48/146 (32%), Positives = 74/146 (49%), Gaps = 7/146 (4%)
Frame = +3
Query: 515 EIAHISNGPYHGNCISIIKPFI--PTQRQMMILQESFTSRVGSYIIYAPSDRQTMDVALR 572
E+ HI+ G HGNC+ ++ M+ILQE++ S ++YAP D Q+++V +
Sbjct: 6 EMVHIAKGQGHGNCVXXLRANAVNANDSSMLILQETWMDASCSVVVYAPVDVQSLNVVMS 185
Query: 573 GEDSKELPILPYGFVV-----CSKSQPNLNAPFGASNNIEDGSLLTLAAQILSTSPHEID 627
G DS + +LP GF + C+ + N G N GSLLT+ QIL S
Sbjct: 186 GGDSAYVALLPSGFAILPDGHCNDNGCNGTLQKGGGGNDGGGSLLTVGFQILVNSLPTAK 365
Query: 628 QVLNVEDITDINTHLATTILNVKDAL 653
L VE + +N ++ TI +K +L
Sbjct: 366 --LTVESVDTVNNLISCTIQKIKASL 437
>BM893519 similar to GP|22475197|gb homeodomain protein GhHOX2 {Gossypium
hirsutum}, partial (13%)
Length = 421
Score = 73.2 bits (178), Expect = 4e-13
Identities = 39/96 (40%), Positives = 56/96 (57%), Gaps = 2/96 (2%)
Frame = +3
Query: 302 IRYCKKFDA-GVWVIADVSFDSSRPNTAP-LSRGWKHPSGCIIREMPHGGCLVTWVEHVE 359
+RYC++ G W I D DS N P R + SGC+I++MP+G VTWVEH +
Sbjct: 6 LRYCQQNAMEGTWAIVDFPVDSFHQNFHPSYPRYCRRSSGCVIQDMPNGYSRVTWVEHAK 185
Query: 360 VEDKIHTHYVYRDLVGNYNLYGAESWIKELQRMCER 395
VE+K H ++ + V + +GA+ W+ LQR CER
Sbjct: 186 VEEK-PVHQIFCNYVYSGMAFGAQRWLGVLQRQCER 290
>TC219552 similar to UP|Q8LJS8 (Q8LJS8) Homeodomain protein GhHOX1, partial
(20%)
Length = 804
Score = 73.2 bits (178), Expect = 4e-13
Identities = 47/153 (30%), Positives = 80/153 (51%), Gaps = 3/153 (1%)
Frame = +1
Query: 504 WDGLSCGNPMHEIAHISNGPYHGNCISIIKPFIPTQRQMMILQESFTSRVGSYIIYAPSD 563
WD +S G + IA+++ G GN ++I + + + ILQ+S+T+ S + YA D
Sbjct: 40 WDIMSSGGTVQSIANLAKGQDRGNAVAI-QTIKSKENSVWILQDSYTNPYESMVAYASVD 216
Query: 564 RQTMDVALRGEDSKELPILPYGFVVCS---KSQPNLNAPFGASNNIEDGSLLTLAAQILS 620
+ + G DS L ILP GF + +S+P + + N E GSL T+A QIL+
Sbjct: 217 ITGIQSVMTGCDSSNLAILPSGFSIIPDGLESRPLVISSRQEEKNTEGGSLFTMAFQILT 396
Query: 621 TSPHEIDQVLNVEDITDINTHLATTILNVKDAL 653
+ L +E + +NT ++ T+ N++ +L
Sbjct: 397 NASPTAK--LTLESVDSVNTLVSCTLRNIRTSL 489
>BG363027
Length = 492
Score = 71.6 bits (174), Expect = 1e-12
Identities = 48/154 (31%), Positives = 76/154 (49%), Gaps = 3/154 (1%)
Frame = +2
Query: 503 QWDGLSCGNPMHEIAHISNGPYHGNCISIIKPFIPTQRQMMILQESFTSRVGSYIIYAPS 562
QWD +S G + +A+++ G GN ++I K + ILQ+S TS S ++YAP
Sbjct: 32 QWDIMSSGGSVQSVANLAKGKDRGNVVNIQK-IQSKDNSVWILQDSCTSAYESMVVYAPV 208
Query: 563 DRQTMDVALRGEDSKELPILPYGFVVCS---KSQPNLNAPFGASNNIEDGSLLTLAAQIL 619
+ + L G DS L ILP GF + + +P + + E GSL T+A QIL
Sbjct: 209 EFAGIQSVLTGCDSSNLAILPSGFSILPDGIEGRPLVISSRQEEKYTEGGSLFTMAFQIL 388
Query: 620 STSPHEIDQVLNVEDITDINTHLATTILNVKDAL 653
+ L E + +N ++ T+ N+K +L
Sbjct: 389 VNPSPTVK--LTTESVESVNNLVSCTLRNIKTSL 484
>BU090220 similar to GP|22475197|gb| homeodomain protein GhHOX2 {Gossypium
hirsutum}, partial (11%)
Length = 267
Score = 68.6 bits (166), Expect = 9e-12
Identities = 35/91 (38%), Positives = 54/91 (58%)
Frame = +2
Query: 335 KHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDLVGNYNLYGAESWIKELQRMCE 394
+ PSGC+I++MP+G VTWVEH +VE+K H ++ + V + +GA+ W+ LQR CE
Sbjct: 5 RRPSGCVIQDMPNGYSRVTWVEHAKVEEK-PVHQIFCNYVYSGMAFGAQRWLGVLQRQCE 181
Query: 395 RSLGSYVEAIPVEETIGVIQTLEGRNSVIKL 425
R I +GVI + R +++KL
Sbjct: 182 RVASLMARNI---SDLGVIPIADARKNLMKL 265
>AW458675 similar to GP|1881536|gb|A A20 {Arabidopsis thaliana}, partial
(11%)
Length = 411
Score = 62.8 bits (151), Expect = 5e-10
Identities = 35/87 (40%), Positives = 53/87 (60%)
Frame = +2
Query: 417 EGRNSVIKLAQRMVKMFCESLTMPGQLELNHLTLASIGGIRVSFRSTTDDDTSQPNGTIV 476
EGR S++KLA+RMV F + L L + +RV R + DD +P+G ++
Sbjct: 11 EGRKSMMKLAERMVLSFSTGVGASTANAWTPLPL-DLENVRVMTRKSVDDP-GRPSGIVL 184
Query: 477 TAATTLWLPLPALKVFEFLKDPTKRSQ 503
+AAT+LWLP+PA +VF+FL+ R+Q
Sbjct: 185 SAATSLWLPVPARRVFDFLRSENTRNQ 265
>BU761633 similar to GP|1814424|gb|A homeodomain protein AHDP {Arabidopsis
thaliana}, partial (12%)
Length = 379
Score = 57.0 bits (136), Expect = 3e-08
Identities = 25/42 (59%), Positives = 31/42 (73%)
Frame = +1
Query: 2 KECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHER 43
KEC HPDE +R L+ + LE KQ+KFWFQN+RT +K Q ER
Sbjct: 253 KECPHPDEKQRLDLSKRLALENKQVKFWFQNRRTQMKTQLER 378
>TC219551 similar to UP|Q8LJS8 (Q8LJS8) Homeodomain protein GhHOX1, partial
(15%)
Length = 671
Score = 52.8 bits (125), Expect = 5e-07
Identities = 35/113 (30%), Positives = 56/113 (48%), Gaps = 3/113 (2%)
Frame = +1
Query: 544 ILQESFTSRVGSYIIYAPSDRQTMDVALRGEDSKELPILPYGFVVCS---KSQPNLNAPF 600
IL +S S ++YA D + G DS L ILP GF + +S+P + +
Sbjct: 13 ILXDSXXXXXESMVVYASVDITXTQSVMTGCDSSNLAILPXGFSIIPDGLESRPLVISSR 192
Query: 601 GASNNIEDGSLLTLAAQILSTSPHEIDQVLNVEDITDINTHLATTILNVKDAL 653
N E GSL T+A QIL+ + L +E + +NT ++ T+ N++ +L
Sbjct: 193 QEEKNTEGGSLFTMAFQILTNASPAAK--LTMESVDSVNTLVSCTLRNIRTSL 345
>BG362843 similar to GP|8885530|dbj| homeodomain transcription factor-like
{Arabidopsis thaliana}, partial (10%)
Length = 466
Score = 50.8 bits (120), Expect = 2e-06
Identities = 32/87 (36%), Positives = 47/87 (53%), Gaps = 6/87 (6%)
Frame = +1
Query: 542 MMILQESFTSRVGSYIIYAPSDRQTMDVALRGEDSKELPILPYGFVVCSKSQPNLNAPFG 601
M+ILQE++ S ++YAP D Q+++V + G DS + +LP GF + N N G
Sbjct: 193 MLILQETWMDASCSVVVYAPVDVQSLNVVMSGGDSAYVALLPSGFAILPDGHCNDNGCNG 372
Query: 602 A------SNNIEDGSLLTLAAQILSTS 622
+ S+ GSLLT+ QIL S
Sbjct: 373 SLQKGRGSDYGSGGSLLTVGFQILVNS 453
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,318,534
Number of Sequences: 63676
Number of extensions: 466060
Number of successful extensions: 2164
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 2137
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2144
length of query: 657
length of database: 12,639,632
effective HSP length: 103
effective length of query: 554
effective length of database: 6,081,004
effective search space: 3368876216
effective search space used: 3368876216
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147010.5


