

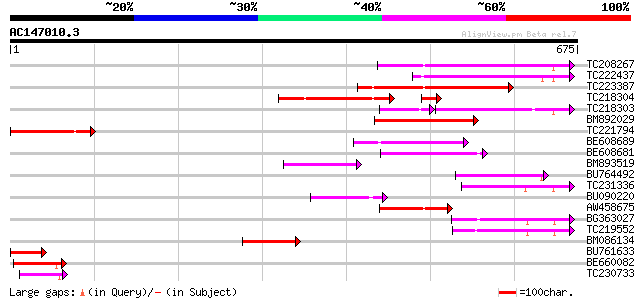
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.3 - phase: 0 /pseudo
(675 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC208267 homologue to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, p... 154 1e-37
TC222437 weakly similar to UP|O23045 (O23045) YUP8H12.16 protein... 150 1e-36
TC223387 141 8e-34
TC218304 similar to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, par... 124 1e-30
TC218303 homologue to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, p... 109 2e-29
BM892029 116 3e-26
TC221794 homologue to UP|Q9FR59 (Q9FR59) Homeobox 1, partial (20%) 97 2e-20
BE608689 similar to GP|3925363|gb| homeodomain protein {Malus x ... 85 9e-17
BE608681 weakly similar to GP|3925363|gb| homeodomain protein {M... 82 1e-15
BM893519 similar to GP|22475197|gb homeodomain protein GhHOX2 {G... 74 3e-13
BU764492 73 4e-13
TC231336 similar to UP|Q9ZTA8 (Q9ZTA8) Homeodomain protein (Frag... 71 2e-12
BU090220 similar to GP|22475197|gb| homeodomain protein GhHOX2 {... 68 2e-11
AW458675 similar to GP|1881536|gb|A A20 {Arabidopsis thaliana}, ... 66 6e-11
BG363027 64 2e-10
TC219552 similar to UP|Q8LJS8 (Q8LJS8) Homeodomain protein GhHOX... 62 8e-10
BM086134 60 3e-09
BU761633 similar to GP|1814424|gb|A homeodomain protein AHDP {Ar... 56 6e-08
BE660082 47 2e-05
TC230733 homologue to UP|Q9SP47 (Q9SP47) Homeodomain-leucine zip... 47 3e-05
>TC208267 homologue to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, partial (31%)
Length = 1049
Score = 154 bits (389), Expect = 1e-37
Identities = 91/240 (37%), Positives = 140/240 (57%), Gaps = 5/240 (2%)
Frame = +2
Query: 438 QTLEGRNSVIKLADRMVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNG 497
QT GR S++KLA+RMV FC ++ L+ VRV R + DD +P G
Sbjct: 35 QTKTGRKSMMKLAERMVISFCAGVSASTAHTWTTLSGTGADDVRVMTRKSADDPG-RPPG 211
Query: 498 TVVTAATTLWLPLPAQKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK 557
V++AAT+ WLP+P ++VF+FL+D R++W+ LS G + E+AHI+NG GNC+S+++
Sbjct: 212 IVLSAATSFWLPVPPKRVFDFLRDENSRNEWDILSNGGVVQEMAHIANGRDTGNCVSLLR 391
Query: 558 --SFIPTQRQMVILQESFTSSVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSK 615
S +Q M+ILQES T+S GS+VIYAP+D M+V L G D + +LP G +
Sbjct: 392 VNSANSSQSNMLILQESCTNSTGSFVIYAPVDIVAMNVVLNGGDPDYVALLPSGFAILPD 571
Query: 616 NQANLNAPFGASKSIEDGSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
+ N G ++ GSL+T+A Q L++ S+ +N+ +A T+ +K +L
Sbjct: 572 GTTSHNGSGGIGETGPSGSLLTVAFQILVDSVPTAKLSLGSVATVNNLIACTVERIKASL 751
>TC222437 weakly similar to UP|O23045 (O23045) YUP8H12.16 protein, partial
(16%)
Length = 719
Score = 150 bits (380), Expect = 1e-36
Identities = 81/205 (39%), Positives = 124/205 (59%), Gaps = 12/205 (5%)
Frame = +1
Query: 480 VRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFEFLKDPTKRSQWNGLSCGNPMHE 539
VRV++ ++D QPNG V++AATT+WLP P VF F KD KR QW+ LS GN + E
Sbjct: 1 VRVTVHKSSDP--GQPNGVVLSAATTIWLPTPPHAVFNFFKDENKRPQWDVLSNGNAVQE 174
Query: 540 IAHISNGPYHGNCISVIKSFIPTQRQMVILQESFTSSVGSYVIYAPIDRKTMDVALRGED 599
+A+I+NG + GN ISV+++F + + M+ILQES S GS+V+Y P+D ++++A+ GED
Sbjct: 175 VANIANGLHPGNSISVLRAFNNSTQNMLILQESCIDSYGSFVVYCPVDLPSINLAMSGED 354
Query: 600 SKELPILPYGLIVCSKNQANLNAPFGASKSIED---------GSLITLAAQTYAI---GQ 647
+P+LP G + Q + GAS S + GSL+T+A Q
Sbjct: 355 PSYIPLLPNGFTILPDGQPDQEGDDGASTSSNNANRNIVRSGGSLVTIAFQILVSSLPSA 534
Query: 648 VLNVDSLNDINSQLASTILNVKDAL 672
LN++S+ +N+ + ST+ +K +L
Sbjct: 535 KLNMESVTTVNNLIGSTVQQIKSSL 609
>TC223387
Length = 569
Score = 141 bits (356), Expect = 8e-34
Identities = 80/187 (42%), Positives = 114/187 (60%), Gaps = 2/187 (1%)
Frame = +2
Query: 415 RMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMFCECLTMPGQVELNHLTL 474
R CER S IP + + VI + EGR S++KLA+RMV +C + L+
Sbjct: 2 RQCERLASSMANNIPAGD-LCVITSAEGRKSMMKLAERMVMSYCTGVGASTAHAWTTLSA 178
Query: 475 DSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFEFLKDPTKRSQWNGLSCG 534
VRV R +TD+ +P G V++AAT+ WLP+P ++VF FL+D R++W+ LS G
Sbjct: 179 TGCDDVRVMTRKSTDEPG-RPPGIVLSAATSFWLPVPPKRVFHFLRDQNSRNEWDILSNG 355
Query: 535 NPMHEIAHISNGPYHGNCISVIK--SFIPTQRQMVILQESFTSSVGSYVIYAPIDRKTMD 592
+ E+AHI+NG GNC+S+++ S +Q M+ILQES T S GSYV+YAP+D M+
Sbjct: 356 GLVQELAHIANGRDPGNCVSLLRVNSANSSQSNMLILQESCTDSTGSYVVYAPVDIVAMN 535
Query: 593 VALRGED 599
V L G D
Sbjct: 536 VVLSGGD 556
>TC218304 similar to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, partial (26%)
Length = 575
Score = 124 bits (311), Expect(2) = 1e-30
Identities = 63/138 (45%), Positives = 87/138 (62%)
Frame = +1
Query: 321 REFNIIRYCKKVDPGVWVITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVE 380
RE +RYCK+ G W + DVS D+ RP+ P +R + PSGC+I+EMP+G V WVE
Sbjct: 7 REIYFVRYCKQHGDGTWAVVDVSLDNLRPS--PSARCRRRPSGCLIQEMPNGYSKVIWVE 180
Query: 381 HVEVEDKIHTHYVYRDLVGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTL 440
HVEV+D+ H +Y+ LV + +GA+ WI L R CER + IP + +GVI
Sbjct: 181 HVEVDDR-GVHNLYKQLVSSGHAFGAKRWIANLDRQCERLASAMATNIPTVD-VGVITNP 354
Query: 441 EGRNSVIKLADRMVKMFC 458
+GR S++KLA+RMV FC
Sbjct: 355 DGRKSMLKLAERMVISFC 408
Score = 27.7 bits (60), Expect(2) = 1e-30
Identities = 10/24 (41%), Positives = 17/24 (70%)
Frame = +2
Query: 491 DASQPNGTVVTAATTLWLPLPAQK 514
D +P G V++AAT+ WLP+ ++
Sbjct: 503 DPGRPPGIVLSAATSFWLPVSPKR 574
>TC218303 homologue to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, partial (32%)
Length = 1230
Score = 109 bits (272), Expect(2) = 2e-29
Identities = 65/171 (38%), Positives = 97/171 (56%), Gaps = 5/171 (2%)
Frame = +2
Query: 507 WLPLPAQKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIK--SFIPTQR 564
WLP+ ++VFEFL+D RS+W+ LS G + E+AHI+NG GNC+S+++ S +Q
Sbjct: 206 WLPVSPKRVFEFLRDENSRSEWDILSNGGVVQEMAHIANGRDTGNCVSLLRVNSANSSQS 385
Query: 565 QMVILQESFTSSVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPF 624
M+ILQES S GS+VIYAP+D M+V L G D + +LP G +
Sbjct: 386 NMLILQESCADSTGSFVIYAPVDIVAMNVVLNGGDPDYVALLPSGFAILPDGTTAHGG-- 559
Query: 625 GASKSIEDGSLITLAAQTYAIG---QVLNVDSLNDINSQLASTILNVKDAL 672
G + GSL+T+A Q L++ S+ +N+ +A T+ +K AL
Sbjct: 560 GIGDTGHGGSLLTVAFQILVDSVPTAKLSLGSVATVNNLIACTVERIKAAL 712
Score = 38.5 bits (88), Expect(2) = 2e-29
Identities = 24/65 (36%), Positives = 36/65 (54%)
Frame = +3
Query: 441 EGRNSVIKLADRMVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVV 500
+GR S++KLA+RMV FC ++ L+ VRV R + DD +P G V+
Sbjct: 12 DGRKSMLKLAERMVISFCAGVSASTAHTWTTLSGTGADDVRVMTRKSV-DDPGRPPGIVL 188
Query: 501 TAATT 505
+AAT+
Sbjct: 189 SAATS 203
>BM892029
Length = 421
Score = 116 bits (291), Expect = 3e-26
Identities = 57/124 (45%), Positives = 78/124 (61%)
Frame = +2
Query: 435 GVIQTLEGRNSVIKLADRMVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQ 494
GVI + EG+ S++KLA RMV FC ++ L+ + V V + D Q
Sbjct: 50 GVIPSPEGKRSMMKLAQRMVTNFCASISSSAGHRWTTLSGSGMNEVGVRVTVHKSSDPGQ 229
Query: 495 PNGTVVTAATTLWLPLPAQKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCIS 554
PNG V++AATT+WLP+P Q VF F KD KR QW+ LS GN + E+AHI+NG + GNCIS
Sbjct: 230 PNGVVLSAATTIWLPIPPQTVFNFFKDEKKRPQWDVLSNGNAVQEVAHIANGSHPGNCIS 409
Query: 555 VIKS 558
V+++
Sbjct: 410 VLRA 421
>TC221794 homologue to UP|Q9FR59 (Q9FR59) Homeobox 1, partial (20%)
Length = 501
Score = 97.1 bits (240), Expect = 2e-20
Identities = 46/101 (45%), Positives = 67/101 (65%)
Frame = +3
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++GLEP Q+K WFQNKR +K QHER N LR EN+K+R +N+
Sbjct: 129 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKTQHERHENTQLRTENEKLRADNM 308
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ +E L C +CGG P + + ++ ENA+L++E
Sbjct: 309 RFREALGNASCPNCGG-PTAIGEMSFDEHHLRLENARLREE 428
>BE608689 similar to GP|3925363|gb| homeodomain protein {Malus x domestica},
partial (18%)
Length = 413
Score = 85.1 bits (209), Expect = 9e-17
Identities = 47/137 (34%), Positives = 75/137 (54%)
Frame = +2
Query: 410 IKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMFCECLTMPGQVEL 469
I LQR CE +I ++ + Q GR S++KLA RM FC + +
Sbjct: 2 IATLQRQCECLAILMSSSISSDDHTALSQA--GRRSMLKLAQRMTSNFCSGVCASSARKW 175
Query: 470 NHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFEFLKDPTKRSQWN 529
+ L + ++G + DD +P G V++AAT++W+P+ Q++F+FL+D RS+W+
Sbjct: 176 DSLHIGTLGDDMKVMTRKNVDDPGEPPGIVLSAATSVWVPVSRQRLFDFLRDERLRSEWD 355
Query: 530 GLSCGNPMHEIAHISNG 546
LS G PM E+ HI+ G
Sbjct: 356 ILSNGGPMQEMVHIAKG 406
>BE608681 weakly similar to GP|3925363|gb| homeodomain protein {Malus x
domestica}, partial (19%)
Length = 475
Score = 81.6 bits (200), Expect = 1e-15
Identities = 40/128 (31%), Positives = 72/128 (56%)
Frame = +2
Query: 442 GRNSVIKLADRMVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVT 501
G S++KLA M +C ++ ++ ++L + ++G + DD +P G V+T
Sbjct: 80 GGKSMLKLAQLMTSNYCSGVSASSALKWDNLHISTLGDDMKVMTRKNVDDPGEPTGIVLT 259
Query: 502 AATTLWLPLPAQKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIKSFIP 561
AAT++W+P+ ++ L D RS+W+ L G PMH++ HI HGNC+S+++ +
Sbjct: 260 AATSVWVPVTRMRLLNSLLDEHLRSEWDILFNGGPMHDMVHIVR*QGHGNCVSILR--LM 433
Query: 562 TQRQMVIL 569
RQ++ L
Sbjct: 434 PLRQLIYL 457
>BM893519 similar to GP|22475197|gb homeodomain protein GhHOX2 {Gossypium
hirsutum}, partial (13%)
Length = 421
Score = 73.6 bits (179), Expect = 3e-13
Identities = 39/96 (40%), Positives = 55/96 (56%), Gaps = 2/96 (2%)
Frame = +3
Query: 326 IRYCKK-VDPGVWVITDVSFDSSRPNTAP-LSRGWKHPSGCIIREMPHGGCLVTWVEHVE 383
+RYC++ G W I D DS N P R + SGC+I++MP+G VTWVEH +
Sbjct: 6 LRYCQQNAMEGTWAIVDFPVDSFHQNFHPSYPRYCRRSSGCVIQDMPNGYSRVTWVEHAK 185
Query: 384 VEDKIHTHYVYRDLVGEYNLYGAESWIKELQRMCER 419
VE+K H ++ + V +GA+ W+ LQR CER
Sbjct: 186 VEEK-PVHQIFCNYVYSGMAFGAQRWLGVLQRQCER 290
>BU764492
Length = 428
Score = 73.2 bits (178), Expect = 4e-13
Identities = 42/122 (34%), Positives = 64/122 (52%), Gaps = 11/122 (9%)
Frame = +3
Query: 531 LSCGNPMHEIAHISNGPYHGNCISVIKSFI--PTQRQMVILQESFTSSVGSYVIYAPIDR 588
LS G PM E+AHI+ G HGN +S++++ Q M+ILQE+ + GS V+YAP+D
Sbjct: 9 LSNGGPMQEMAHIAKGQDHGNAVSLLRASAINSNQSSMLILQETCIDAAGSLVVYAPVDI 188
Query: 589 KTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGASKSIE---------DGSLITLA 639
+ V + G DS + +LP G + + P G + + GSL+T+A
Sbjct: 189 PVLYVVMNGGDSAYVALLPSGFAIVPDGPGSRGPPNGPTSTTNGGDNGVTRVSGSLLTVA 368
Query: 640 AQ 641
Q
Sbjct: 369 FQ 374
>TC231336 similar to UP|Q9ZTA8 (Q9ZTA8) Homeodomain protein (Fragment),
partial (19%)
Length = 869
Score = 70.9 bits (172), Expect = 2e-12
Identities = 43/144 (29%), Positives = 76/144 (51%), Gaps = 10/144 (6%)
Frame = +3
Query: 539 EIAHISNGPYHGNCISVIKSFI--PTQRQMVILQESFTSSVGSYVIYAPIDRKTMDVALR 596
E+ HI+ G HGNC+ +++ M+ILQE++ + S V+YAP+D ++++V +
Sbjct: 6 EMVHIAKGQGHGNCVXXLRANAVNANDSSMLILQETWMDASCSVVVYAPVDVQSLNVVMS 185
Query: 597 GEDSKELPILPYGLIV-----CSKNQANLNAPFGASKSIEDGSLITLAAQTYAIG---QV 648
G DS + +LP G + C+ N N G + GSL+T+ Q
Sbjct: 186 GGDSAYVALLPSGFAILPDGHCNDNGCNGTLQKGGGGNDGGGSLLTVGFQILVNSLPTAK 365
Query: 649 LNVDSLNDINSQLASTILNVKDAL 672
L V+S++ +N+ ++ TI +K +L
Sbjct: 366 LTVESVDTVNNLISCTIQKIKASL 437
>BU090220 similar to GP|22475197|gb| homeodomain protein GhHOX2 {Gossypium
hirsutum}, partial (11%)
Length = 267
Score = 67.8 bits (164), Expect = 2e-11
Identities = 35/91 (38%), Positives = 53/91 (57%)
Frame = +2
Query: 359 KHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDLVGEYNLYGAESWIKELQRMCE 418
+ PSGC+I++MP+G VTWVEH +VE+K H ++ + V +GA+ W+ LQR CE
Sbjct: 5 RRPSGCVIQDMPNGYSRVTWVEHAKVEEK-PVHQIFCNYVYSGMAFGAQRWLGVLQRQCE 181
Query: 419 RSLGSNVEAIPVEETIGVIQTLEGRNSVIKL 449
R I +GVI + R +++KL
Sbjct: 182 RVASLMARNI---SDLGVIPIADARKNLMKL 265
>AW458675 similar to GP|1881536|gb|A A20 {Arabidopsis thaliana}, partial
(11%)
Length = 411
Score = 65.9 bits (159), Expect = 6e-11
Identities = 38/87 (43%), Positives = 55/87 (62%)
Frame = +2
Query: 441 EGRNSVIKLADRMVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVV 500
EGR S++KLA+RMV F + L LD + VRV R + DD +P+G V+
Sbjct: 11 EGRKSMMKLAERMVLSFSTGVGASTANAWTPLPLD-LENVRVMTRKSVDDPG-RPSGIVL 184
Query: 501 TAATTLWLPLPAQKVFEFLKDPTKRSQ 527
+AAT+LWLP+PA++VF+FL+ R+Q
Sbjct: 185 SAATSLWLPVPARRVFDFLRSENTRNQ 265
>BG363027
Length = 492
Score = 63.9 bits (154), Expect = 2e-10
Identities = 46/153 (30%), Positives = 79/153 (51%), Gaps = 7/153 (4%)
Frame = +2
Query: 527 QWNGLSCGNPMHEIAHISNGPYHGNCISVIKSFIPTQRQMV-ILQESFTSSVGSYVIYAP 585
QW+ +S G + +A+++ G GN +++ K I ++ V ILQ+S TS+ S V+YAP
Sbjct: 32 QWDIMSSGGSVQSVANLAKGKDRGNVVNIQK--IQSKDNSVWILQDSCTSAYESMVVYAP 205
Query: 586 IDRKTMDVALRGEDSKELPILPYGLIVCS---KNQANLNAPFGASKSIEDGSLITLAAQT 642
++ + L G DS L ILP G + + + + + K E GSL T+A Q
Sbjct: 206 VEFAGIQSVLTGCDSSNLAILPSGFSILPDGIEGRPLVISSRQEEKYTEGGSLFTMAFQI 385
Query: 643 YAIGQ---VLNVDSLNDINSQLASTILNVKDAL 672
L +S+ +N+ ++ T+ N+K +L
Sbjct: 386 LVNPSPTVKLTTESVESVNNLVSCTLRNIKTSL 484
>TC219552 similar to UP|Q8LJS8 (Q8LJS8) Homeodomain protein GhHOX1, partial
(20%)
Length = 804
Score = 62.0 bits (149), Expect = 8e-10
Identities = 41/151 (27%), Positives = 79/151 (52%), Gaps = 6/151 (3%)
Frame = +1
Query: 528 WNGLSCGNPMHEIAHISNGPYHGNCISVIKSFIPTQRQMVILQESFTSSVGSYVIYAPID 587
W+ +S G + IA+++ G GN ++ I++ + + ILQ+S+T+ S V YA +D
Sbjct: 40 WDIMSSGGTVQSIANLAKGQDRGNAVA-IQTIKSKENSVWILQDSYTNPYESMVAYASVD 216
Query: 588 RKTMDVALRGEDSKELPILPYGLIVCS---KNQANLNAPFGASKSIEDGSLITLAAQTYA 644
+ + G DS L ILP G + +++ + + K+ E GSL T+A Q
Sbjct: 217 ITGIQSVMTGCDSSNLAILPSGFSIIPDGLESRPLVISSRQEEKNTEGGSLFTMAFQILT 396
Query: 645 IGQ---VLNVDSLNDINSQLASTILNVKDAL 672
L ++S++ +N+ ++ T+ N++ +L
Sbjct: 397 NASPTAKLTLESVDSVNTLVSCTLRNIRTSL 489
>BM086134
Length = 428
Score = 60.1 bits (144), Expect = 3e-09
Identities = 28/69 (40%), Positives = 43/69 (61%)
Frame = +1
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F IV++A T++V G G+ +GAL +M E+ + +PLV RE +RYCK+ G W
Sbjct: 217 VFSGIVSRAMTLEVLSTGVAGNYNGALQVMTAELQLPTPLVPTRESYFVRYCKQHADGTW 396
Query: 338 VITDVSFDS 346
+ DVS D+
Sbjct: 397 AVVDVSLDN 423
>BU761633 similar to GP|1814424|gb|A homeodomain protein AHDP {Arabidopsis
thaliana}, partial (12%)
Length = 379
Score = 55.8 bits (133), Expect = 6e-08
Identities = 24/42 (57%), Positives = 30/42 (71%)
Frame = +1
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHER 43
KEC HPDE QR L+ ++ LE KQ+K WFQN+R +K Q ER
Sbjct: 253 KECPHPDEKQRLDLSKRLALENKQVKFWFQNRRTQMKTQLER 378
>BE660082
Length = 846
Score = 47.4 bits (111), Expect = 2e-05
Identities = 22/70 (31%), Positives = 43/70 (61%), Gaps = 7/70 (10%)
Frame = +2
Query: 5 HHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRREND-------KIR 57
H + ++ +LA+++GL+P+Q+ WFQN+RA K++ E L++ ++ ++
Sbjct: 326 HKLESERKDRLAMELGLDPRQVAVWFQNRRARWKNKKLEEEYSNLKKNHEATLLEKCRLE 505
Query: 58 NENLKIKEEL 67
E LK+KE+L
Sbjct: 506 TEVLKLKEQL 535
>TC230733 homologue to UP|Q9SP47 (Q9SP47) Homeodomain-leucine zipper
protein 57, partial (76%)
Length = 1305
Score = 47.0 bits (110), Expect = 3e-05
Identities = 23/64 (35%), Positives = 38/64 (58%), Gaps = 7/64 (10%)
Frame = +3
Query: 12 RCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIR-------NENLKIK 64
+ QLA ++GL+P+Q+ WFQN+RA K + + G L+ D+++ EN K+K
Sbjct: 3 KVQLAKELGLQPRQVAIWFQNRRARFKTKQLEKDYGVLKASYDRLKGDYESLVQENDKLK 182
Query: 65 EELK 68
E+K
Sbjct: 183AEVK 194
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.330 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,195,464
Number of Sequences: 63676
Number of extensions: 508375
Number of successful extensions: 2970
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 2939
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2954
length of query: 675
length of database: 12,639,632
effective HSP length: 104
effective length of query: 571
effective length of database: 6,017,328
effective search space: 3435894288
effective search space used: 3435894288
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147010.3


