

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.11 - phase: 0 /pseudo
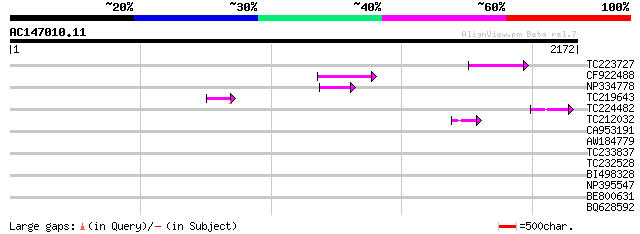
(2172 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 104 5e-22
CF922488 99 2e-20
NP334778 reverse transcriptase [Glycine max] 76 2e-13
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 65 3e-10
TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 47 9e-05
TC212032 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 45 5e-04
CA953191 43 0.001
AW184779 43 0.001
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 40 0.011
TC232528 weakly similar to UP|Q6WAY5 (Q6WAY5) Gag/pol polyprotei... 39 0.025
BI498328 38 0.042
NP395547 reverse transcriptase [Glycine max] 37 0.072
BE800631 weakly similar to GP|9294065|dbj| contains similarity t... 33 1.4
BQ628592 26 1.4
>TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (9%)
Length = 843
Score = 104 bits (259), Expect = 5e-22
Identities = 90/233 (38%), Positives = 107/233 (45%)
Frame = +2
Query: 1756 TIDPGIMTSNSSC*AVSIRQVLPNKIRRP*EDWPVDSC*MEIFCTRETMTWYC*DVLMNM 1815
T+ GI TS+ *A + Q LP IR *E W S * E +C RET T V M
Sbjct: 128 TVSLGISTSSDMS*AKNTCQRLPTMIRGH*EGWRPVSS*AEAYCIRETTT*NLCGVWMPG 307
Query: 1816 KQSS*CMTYMTVPSGPMLQGILCQGSCYEQVTTGWPWSMIATSTPENATNVKSMLIRFMC 1875
+Q +* M G+L G EQV TG PW +I S NATNVK I M
Sbjct: 308 RQIT*SRKSMRARLERTPTGMLWPGRS*EQVITGLPWKVIVVSM*GNATNVKRSQIMSMP 487
Query: 1876 LHTLSILCHPHGRSQCGAST*LEELNRRLQMVIVSS*WQLTTSPNGLKQHLIPM*PSKW* 1935
L + C P G S CG L+ R +MVI SS + SP+G +Q IP W
Sbjct: 488 HRIL*MSCPPLGLSPCGE*MSSGPLSPRPRMVIASSS*R*IISPSGSRQLPIPTS*GVWW 667
Query: 1936 PSSSRTTSSVDMVFPARLLLTMVPT*TTMWCKLFVKNSKLSIITLLPIDLR*M 1988
S R SS DMV RL T PT* T F ++ K SI P R*+
Sbjct: 668 SGSLRKRSSADMVCQGRLSRTTAPT*ITR*WGKFARSLKSSITIPXPTGQR*I 826
>CF922488
Length = 741
Score = 99.0 bits (245), Expect = 2e-20
Identities = 89/227 (39%), Positives = 112/227 (49%)
Frame = +1
Query: 1179 RRMVKSGCVLTSET*TKPVQKTTFRYLILMCLLITLLSLRCSPSWTVSPVTIRSRCLLKI 1238
+RM + C T E *TKPVQ+ +F Y I IT SPSW V R R +I
Sbjct: 10 KRMGRCECAWTIEI*TKPVQRISFLYRISTFSWITRPVFPNSPSWMDFQVITR*R*HQRI 189
Query: 1239 EKRRLLSLHGVPSATK*CRSA*SMLVLPTKGE*LLCFMT*FTKKSKYMWTT*L*NQQMRS 1298
KR+L L+G PSA + CR * ML T G T* T++ + W T* *NQ+ R
Sbjct: 190 WKRQLSLLYGEPSAIRLCRLG*RMLGQHTSGPWWHYSRT*CTRR*RSTWMT*S*NQERRR 369
Query: 1299 SMLNIWQRCLKG*ENTSFD*IPTNVHSASDPGSY*ASLSAKRALKSILIKSGPSEKCQLH 1358
+ L+I + CL NT D*IP +V +P S L A+ + I + S + H
Sbjct: 370 NTLSICESCLGDYVNTG*D*IPQSVCLR*NPESCSTLLIAREE*RWIRTR*K*SLRWPSH 549
Query: 1359 RQRSKSEASSDV*ITSPDSYLT*PQPAGRSSSYSGRISLLYGMMNAK 1405
QRSKS+ S * TS DSY +* A S RISL G M K
Sbjct: 550 IQRSKSKVSWGG*TTS*DSYHS*LPLASLFSYCCARISLSNGTMIVK 690
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 75.9 bits (185), Expect = 2e-13
Identities = 59/141 (41%), Positives = 75/141 (52%)
Frame = +1
Query: 1185 GCVLTSET*TKPVQKTTFRYLILMCLLITLLSLRCSPSWTVSPVTIRSRCLLKIEKRRLL 1244
GCV T E *T+PVQ+ F Y +T L S SW V IR R +I KR+L
Sbjct: 4 GCVWTIEI*TEPVQRIIFLYRTSTFSWLTWPVLPYSLSWMDFRVIIR*RWHQRIWKRQLS 183
Query: 1245 SLHGVPSATK*CRSA*SMLVLPTKGE*LLCFMT*FTKKSKYMWTT*L*NQQMRSSMLNIW 1304
L+G PSA +*CR * +L PT G T* T++S+ MW * NQ+ R + +I
Sbjct: 184 LLYGEPSAIR*CRLG*RILGQPTIGPWWHYSRT*CTRRSRPMWMK*SRNQEWRRNT*SIC 363
Query: 1305 QRCLKG*ENTSFD*IPTNVHS 1325
+ CL NT D*IP +V S
Sbjct: 364 KICLGNYVNTG*D*IPGSVCS 426
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein
(Fragment), partial (8%)
Length = 1320
Score = 65.1 bits (157), Expect = 3e-10
Identities = 46/125 (36%), Positives = 58/125 (45%), Gaps = 15/125 (12%)
Frame = +1
Query: 755 PWIHDAGAVTSTLHQKLKFIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAE-GTAFQGLT 813
PWIH G V STLHQKLKF+ G LV V GEE LVS SS +EA TAFQ
Sbjct: 1 PWIHSVGVVPSTLHQKLKFVVEGHLVIVSGEEDILVSCPSSMPYVEAAEESLETAFQSFE 180
Query: 814 IEGAEPKKAGAAMASLKD-----AQKVIQDGQTAGWG---------KVIQLCENKRKEGL 859
+ + L D A+ ++ +G G G +I N+ K GL
Sbjct: 181 VVSISSVDSLFGQPCLSDAAVMMARVMLGNGYEPGMGLGKDNGGITSLINTQGNRGKYGL 360
Query: 860 GFSPS 864
G+ P+
Sbjct: 361 GYKPT 375
>TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 669
Score = 47.0 bits (110), Expect = 9e-05
Identities = 53/165 (32%), Positives = 76/165 (45%), Gaps = 1/165 (0%)
Frame = +2
Query: 1994 PTRISRELSRRW*PLTRTGMRCYPMLCMATVLQCAVRPGQPLSLLYMVWKQFFLWKWRSH 2053
P RIS+ LS+R TR G RC T LQC + GQ S YM W+ + + +S
Sbjct: 8 PIRISKRLSKR*PCHTRIGTRCSHSRYTVTGLQCERQLGQRRSHWYMGWRLCYRLR*KSR 187
Query: 2054 PSV*SWKQSYLR-LNGAKAGTIS*I*LRKNVWMPWLVDSHIKQK*RLLLTRKSILENSR* 2112
SW+ R +G K IS LR + P ++ + ++*R+ TR+ +S
Sbjct: 188 -H*GSWQNPD*RNQSGLKRAMISSTSLRVSA*RP*VMGACTSKE*RVHSTRRYACASSMR 364
Query: 2113 GNLY*KGG*ASNPTQGASGRLTTKVLMLSRRPSPVVL*SLHTWMM 2157
L *+ + T +G TTK L+L R P L TWM+
Sbjct: 365 ETLC*RKCPMLSRTIEGNGPRTTKGLLL*RGLFPEEPWCLPTWMV 499
>TC212032 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (3%)
Length = 803
Score = 44.7 bits (104), Expect = 5e-04
Identities = 35/117 (29%), Positives = 49/117 (40%)
Frame = +3
Query: 1692 LQRLSCTIFLVMRTKWLMLLLLCPPCFE*TIGMMCQ*SKCNASKDLRMCLLLGM*SIRLV 1751
L R L + KW M L L PC * + + L + ++ +
Sbjct: 354 LMRSPSITLLERKIKWQMRLPL*CPC--------SS*HRMGTYRTLSLGVVADPHIVVWW 509
Query: 1752 KMWLTIDPGIMTSNSSC*AVSIRQVLPNKIRRP*EDWPVDSC*MEIFCTRETMTWYC 1808
K T+ GI+ S+ + A S LP + DW S * E +CTRETMTW+C
Sbjct: 510 KRNGTVSLGILISSDTLKAKSTHWRLPTTTKGERGDWQPASS*AEAYCTRETMTWFC 680
>CA953191
Length = 422
Score = 43.1 bits (100), Expect = 0.001
Identities = 27/58 (46%), Positives = 34/58 (58%), Gaps = 1/58 (1%)
Frame = -3
Query: 744 INASYSCLLGRPW-IHDAGAVTSTLHQKLKFIRNGKLVTVHGEEAYLVSQLSSFSCIE 800
I +Y+ L GRPW IH V STLH K K + +GKLV + +E LV + SS IE
Sbjct: 420 ITPTYNGLQGRPWRIHCVKLVPSTLH*K*KIVIDGKLVIIFVKEDLLVGEPSSTPYIE 247
>AW184779
Length = 432
Score = 43.1 bits (100), Expect = 0.001
Identities = 27/45 (60%), Positives = 30/45 (66%)
Frame = +1
Query: 1891 CGAST*LEELNRRLQMVIVSS*WQLTTSPNGLKQHLIPM*PSKW* 1935
CGA T* E L+ RLQM I S * QLTTSPNG KQ + +* W*
Sbjct: 292 CGA*T*SEPLSPRLQMDITSF*SQLTTSPNGSKQFRMLV*LGVW* 426
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 40.0 bits (92), Expect = 0.011
Identities = 37/109 (33%), Positives = 53/109 (47%)
Frame = +3
Query: 1223 WTVSPVTIRSRCLLKIEKRRLLSLHGVPSATK*CRSA*SMLVLPTKGE*LLCFMT*FTKK 1282
W VS I+ R +K+ +R L S +G SA +* S * +L P C M * +K
Sbjct: 12 WMVSRGIIKYRWHVKM*RRPLSSPYGGHSAIE*WPSG*KILGQPISVPWWRCSMI*CIRK 191
Query: 1283 SKYMWTT*L*NQQMRSSMLNIWQRCLKG*ENTSFD*IPTNVHSASDPGS 1331
+ T*L + +R + L+I CL+G NT+ +* N H GS
Sbjct: 192 *RST*MT*LPSLGLRPNTLSICVSCLEGCRNTN*N*TQPNAHLG*SRGS 338
>TC232528 weakly similar to UP|Q6WAY5 (Q6WAY5) Gag/pol polyprotein
(Fragment), partial (3%)
Length = 449
Score = 38.9 bits (89), Expect = 0.025
Identities = 16/23 (69%), Positives = 20/23 (86%)
Frame = +1
Query: 710 VKAFDGSRKNVLGEIDLPITIGP 732
V+AFDG+R+ V GEIDLP+ IGP
Sbjct: 373 VRAFDGTRREVRGEIDLPVQIGP 441
>BI498328
Length = 335
Score = 38.1 bits (87), Expect = 0.042
Identities = 24/57 (42%), Positives = 29/57 (50%)
Frame = +2
Query: 1591 SGV*SLMVLSMLMVKELGQSLYPHRGITFLLPPEFCSNVQTIWPSMKHVSLGSRKQL 1647
+G+ + M ML E GQSLYP FL + V TIWPS KH G R+ L
Sbjct: 149 NGLFASMGHPMLWATE*GQSLYPRMISVFLSRLD*VLIVPTIWPSTKHAPSGFRRPL 319
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 37.4 bits (85), Expect = 0.072
Identities = 32/102 (31%), Positives = 48/102 (46%)
Frame = +2
Query: 1186 CVLTSET*TKPVQKTTFRYLILMCLLITLLSLRCSPSWTVSPVTIRSRCLLKIEKRRLLS 1245
CVL + KP +KT + + L L + SWT + VTIR + +L+I+K++LL
Sbjct: 167 CVLIIGSSMKPQEKTITHFPSWIKCLRDLQGNPSTVSWTDTQVTIRLQWILRIKKKQLLH 346
Query: 1246 LHGVPSATK*CRSA*SMLVLPTKGE*LLCFMT*FTKKSKYMW 1287
+ V CRS M +L + * MT K +W
Sbjct: 347 VLSVFLLIAACRSVYVMPLLLFRDV*WQFLMTW*RNVLKSLW 472
>BE800631 weakly similar to GP|9294065|dbj| contains similarity to myb
proteins~gene_id:MRC8.8 {Arabidopsis thaliana}, partial
(8%)
Length = 413
Score = 33.1 bits (74), Expect = 1.4
Identities = 16/37 (43%), Positives = 22/37 (59%)
Frame = +3
Query: 1514 LLLLERLHAGRCFCLNMILCSKLKRQSKVAFLPIILL 1550
+L+L GRC+ LNM LC K + +SK L + LL
Sbjct: 117 VLILNAGRDGRCYVLNMFLCFKKQERSKGLLLVVTLL 227
>BQ628592
Length = 423
Score = 26.2 bits (56), Expect(2) = 1.4
Identities = 24/64 (37%), Positives = 31/64 (47%), Gaps = 1/64 (1%)
Frame = -2
Query: 1404 AKKLLIASRIT-CWNHLSLSHPWKEGL*LCIWQCLMNPWDVYLVNKMKLGRKSMLSTI*A 1462
+KK ASRI C HL +E L C C + WD + LG+++ TI*A
Sbjct: 374 SKKSNRASRIPRCSCHL*-----QEDLFSCT*LC*TSLWDACWFSTTTLGKRNKPFTI*A 210
Query: 1463 RSSP 1466
RS P
Sbjct: 209 RSLP 198
Score = 25.4 bits (54), Expect(2) = 1.4
Identities = 14/38 (36%), Positives = 20/38 (51%)
Frame = -1
Query: 1495 IIQLG*YPEWIRSSISLRKLLLLERLHAGRCFCLNMIL 1532
+I G +P+WI + SLR + GR + LN IL
Sbjct: 114 VIPRGLFPKWIL*NTSLRSRPSRDESLGGRYYYLNSIL 1
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.354 0.154 0.541
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 106,096,087
Number of Sequences: 63676
Number of extensions: 1681120
Number of successful extensions: 21142
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 11411
Number of HSP's successfully gapped in prelim test: 842
Number of HSP's that attempted gapping in prelim test: 8892
Number of HSP's gapped (non-prelim): 13897
length of query: 2172
length of database: 12,639,632
effective HSP length: 112
effective length of query: 2060
effective length of database: 5,507,920
effective search space: 11346315200
effective search space used: 11346315200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147010.11


