

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147007.4 - phase: 0
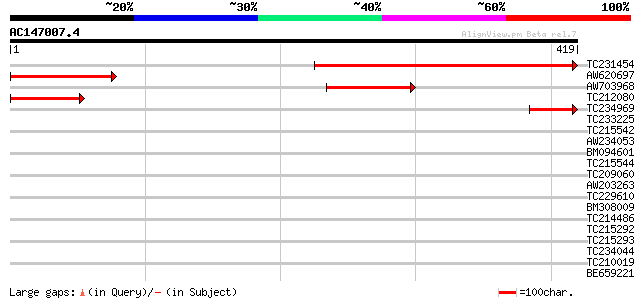
(419 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC231454 weakly similar to UP|Q9ZW88 (Q9ZW88) F5A8.10 protein, p... 269 2e-72
AW620697 110 2e-24
AW703968 96 2e-20
TC212080 similar to UP|Q9ZW88 (Q9ZW88) F5A8.10 protein, partial ... 80 1e-15
TC234969 64 1e-10
TC233225 40 0.001
TC215542 35 0.063
AW234053 35 0.082
BM094601 34 0.14
TC215544 33 0.18
TC209060 weakly similar to GB|AAP42728.1|30984530|BT008715 At4g0... 33 0.31
AW203263 32 0.41
TC229610 weakly similar to UP|O22512 (O22512) Grr1, partial (52%) 32 0.70
BM308009 similar to PIR|D86241|D862 protein T16B5.8 [imported] -... 31 0.91
TC214486 31 1.2
TC215292 similar to UP|FENR_PEA (P10933) Ferredoxin--NADP reduct... 31 1.2
TC215293 similar to UP|FENR_TOBAC (O04977) Ferredoxin--NADP redu... 30 1.6
TC234044 30 2.0
TC210019 similar to UP|Q84KK5 (Q84KK5) S-adenosyl-L-methionine: ... 30 2.6
BE659221 homologue to PIR|AB0167|AB01 ATP-dependent Clp proteina... 30 2.6
>TC231454 weakly similar to UP|Q9ZW88 (Q9ZW88) F5A8.10 protein, partial (46%)
Length = 696
Score = 269 bits (687), Expect = 2e-72
Identities = 125/194 (64%), Positives = 155/194 (79%)
Frame = +2
Query: 226 HLKDCSFDAFELINKGTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMIS 285
H KDC+ + FEL KG LK K+DDVSVIHLDIG+ ENLE VD+ NF +W FY MIS
Sbjct: 2 HXKDCALEFFELXXKGXLKHFKIDDVSVIHLDIGETVENLETVDISNFPIIWPKFYQMIS 181
Query: 286 KASKLKKLRLWSVVFDDEDEVVDIETISVCFPRLTHLSLSYDLKDGVLHYGLQGLSFLMN 345
++S LK+LRLW V+FDDEDEVVD+ETI+ CFP L+HLSLSYD++DGVL+YGLQG S+L +
Sbjct: 182 RSSNLKRLRLWDVMFDDEDEVVDLETIATCFPYLSHLSLSYDVRDGVLYYGLQGSSYLES 361
Query: 346 VVVLELGWTTISDLFSVWVAGLLEGCPNLKKMVIYGYVAEIKTHEECQTFTKFTEFMIQL 405
VVVLELGWT I+DLFS WV GLL+ CPNLKK+VI+G V+E K+HEECQ FT M++L
Sbjct: 362 VVVLELGWTVINDLFSHWVEGLLKRCPNLKKLVIHGIVSEAKSHEECQMLANFTTSMVEL 541
Query: 406 GRKYSHIKFEFEYE 419
R+Y+H+ F+YE
Sbjct: 542 MRRYTHVDPYFKYE 583
>AW620697
Length = 415
Score = 110 bits (274), Expect = 2e-24
Identities = 52/79 (65%), Positives = 63/79 (78%)
Frame = +3
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
ME LPVEV+G ILS LGSARDV++AS+TCK WREAW+ HL +LSFN DWPL+ + S+
Sbjct: 177 MEHLPVEVIGNILSLLGSARDVVVASVTCKTWREAWRNHLHSLSFNYCDWPLFHDSTSAR 356
Query: 61 LEMLITCTLFQTKGLQCLT 79
LE+LIT T+FQT LQ LT
Sbjct: 357 LEILITQTIFQTNALQSLT 413
>AW703968
Length = 206
Score = 96.3 bits (238), Expect = 2e-20
Identities = 44/66 (66%), Positives = 52/66 (78%)
Frame = +2
Query: 235 FELINKGTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLR 294
FELI KGTLK K+DDVSVIHLDIG+ ENLE VD+ NF +W FY MIS++S LK+LR
Sbjct: 8 FELIGKGTLKHFKIDDVSVIHLDIGETVENLETVDISNFTIIWPKFYQMISRSSNLKRLR 187
Query: 295 LWSVVF 300
LW V+F
Sbjct: 188 LWDVMF 205
>TC212080 similar to UP|Q9ZW88 (Q9ZW88) F5A8.10 protein, partial (13%)
Length = 628
Score = 80.5 bits (197), Expect = 1e-15
Identities = 36/55 (65%), Positives = 45/55 (81%)
Frame = +1
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYRE 55
M+ LPVEV+G ILSHL +ARDV+IAS +C+KWR A HL TLSFN+ DWP+YR+
Sbjct: 463 MDQLPVEVIGNILSHLRAARDVVIASASCRKWRLACCKHLHTLSFNSKDWPIYRD 627
>TC234969
Length = 364
Score = 63.9 bits (154), Expect = 1e-10
Identities = 25/35 (71%), Positives = 31/35 (88%)
Frame = +1
Query: 385 EIKTHEECQTFTKFTEFMIQLGRKYSHIKFEFEYE 419
E+KTHEECQ +F+EF++QLGRKY H+KFEFEYE
Sbjct: 13 EVKTHEECQILARFSEFILQLGRKYGHVKFEFEYE 117
>TC233225
Length = 888
Score = 40.4 bits (93), Expect = 0.001
Identities = 57/257 (22%), Positives = 115/257 (44%), Gaps = 17/257 (6%)
Frame = +1
Query: 86 HEFSVT--------PVIAWLMY-TRDSLRELRYNVRTSPNFNI---IEKCSRKT-LEVLT 132
H+F +T + W+++ +R+ ++EL + F I + C + T L++
Sbjct: 10 HKFQITNSKLQSCPEIDQWILFLSRNDIKELVMELGEGEFFRIPSSLFNCGKLTRLDLSR 189
Query: 133 LASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSACPKLETLSIVCPEIAMSDSE 192
+P + F CL+SL+L V IS + L+S CP LE+LS ++ D+
Sbjct: 190 CEFDP----PHSFKGFVCLRSLNLHQVLISPDAIESLISRCPLLESLS-----LSYFDNL 342
Query: 193 ASIELSSSSLKDFFVESYSFDKLI----LVADMLENLHLKDCSFDAFELINKGTLKVLKL 248
A + + + +LK ++E D + L+ ++ +++ D + FE I+ V L
Sbjct: 343 A-LTICAPNLKYLYLEGEFKDICLEDTPLLVEITIAMYMTDDIAEHFEQISNCNF-VKFL 516
Query: 249 DDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLRLWSVVFDDEDEVVD 308
V + +G ++ ++ + +F + + L+ + L+ V F+D E++
Sbjct: 517 GGVPNLEKLVG-------LIYFTKYLSIGIDFVHPPMMYNNLETIELYQVNFEDMVEILV 675
Query: 309 IETISVCFPRLTHLSLS 325
I + P L L +S
Sbjct: 676 ILRLITSSPNLKELQIS 726
>TC215542
Length = 1695
Score = 35.0 bits (79), Expect = 0.063
Identities = 37/132 (28%), Positives = 54/132 (40%), Gaps = 14/132 (10%)
Frame = +3
Query: 2 EDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSHL 61
+D+PVE++ ILS L + V+IAS C+ WR+A L LS + W CS ++
Sbjct: 231 KDIPVELLMQILS-LVDDQTVIIASGVCRGWRDAIYFGLARLSLS---W------CSKNM 380
Query: 62 EMLITCTLFQTKGLQCLTIFMDDE--------------HEFSVTPVIAWLMYTRDSLREL 107
L+ + + LQ L + D HE + + T SL EL
Sbjct: 381 NNLVLSLVPKFAKLQTLILRQDKPQLEDNAVETIAKCCHELQILDLSKSFKLTDRSLYEL 560
Query: 108 RYNVRTSPNFNI 119
R NI
Sbjct: 561 ALGCRDLTKLNI 596
>AW234053
Length = 411
Score = 34.7 bits (78), Expect = 0.082
Identities = 28/103 (27%), Positives = 48/103 (46%), Gaps = 15/103 (14%)
Frame = +3
Query: 226 HLKDCSFDAFELINK--GTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYN- 282
HL + D F+L N G + L DVS+ + DN +E++ + +N
Sbjct: 99 HLLGDNLDLFQLKNGLGGLTSLQTLCDVSI---PVDDNDNGVELIRKLGKLKQLRNLSLN 269
Query: 283 ------------MISKASKLKKLRLWSVVFDDEDEVVDIETIS 313
+++ + L+KL +WS +DEDE++D+ TIS
Sbjct: 270 GVKEEQGSILCFSLNEMTNLEKLNIWS---EDEDEIIDLPTIS 389
>BM094601
Length = 429
Score = 33.9 bits (76), Expect = 0.14
Identities = 31/100 (31%), Positives = 46/100 (46%), Gaps = 11/100 (11%)
Frame = +3
Query: 96 WLMYTRDSLRELRYNVRTSPNFNIIEKCS-----RKTLEVLTLA-SNPISGVEPKY---- 145
WL R S L + T+P+F++I S L L+L+ S P+ + P
Sbjct: 102 WLRLHRSSTTSLSLRL-TTPHFSLIPSLSSFLSHHPFLSSLSLSLSPPLFSLSPHLLSLI 278
Query: 146 -HKFPCLKSLSLSFVSISALDLSLLLSACPKLETLSIVCP 184
F L SLSL+ +S L L ++CP+L +L I P
Sbjct: 279 ISPFSNLLSLSLTPAPVSLSSLLSLSASCPRLNSLRITLP 398
>TC215544
Length = 1377
Score = 33.5 bits (75), Expect = 0.18
Identities = 28/82 (34%), Positives = 41/82 (49%)
Frame = +3
Query: 2 EDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSHL 61
+D+PVE++ ILS L + V+IAS C+ WREA L LS + W CS ++
Sbjct: 234 KDIPVELLMQILS-LVDDQTVIIASEVCRGWREAICLGLTRLSLS---W------CSKNM 383
Query: 62 EMLITCTLFQTKGLQCLTIFMD 83
L+ + LQ L + D
Sbjct: 384 NNLVLSLSPKFTKLQTLILRQD 449
>TC209060 weakly similar to GB|AAP42728.1|30984530|BT008715 At4g08980
{Arabidopsis thaliana;} , partial (64%)
Length = 901
Score = 32.7 bits (73), Expect = 0.31
Identities = 49/212 (23%), Positives = 79/212 (37%), Gaps = 35/212 (16%)
Frame = +3
Query: 3 DLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWP--LYREICSSH 60
+L + +G+I ++L V + CK W A + DW +
Sbjct: 273 ELIPDALGVIFTNLSLQERVTVIPRVCKSWANAVTGPYCWQEIDIKDWSNRCQPDQLDRL 452
Query: 61 LEMLITCTL-----FQTKGLQCLTIF-------------------MDDEHEFSVTPVIAW 96
LEMLIT + GLQ +IF M+D S+ IA
Sbjct: 453 LEMLITRSCGTLRKLSVSGLQTESIFTFIAENACSLHTLRLPRSSMND----SIVEQIAG 620
Query: 97 LMYTRDSLRELRYNVRTSPNFNIIEKCSRKTLEVLTLASNPISGVEPKYHK--------- 147
+ + S ++ Y ++ P + + K LE L +P+ E +
Sbjct: 621 RL-SMISFLDVSYCIKIGPYALEMIGKNCKLLEGLCRNMHPLDTAEKPHQDDEAYAMAST 797
Query: 148 FPCLKSLSLSFVSISALDLSLLLSACPKLETL 179
P LK L +++ IS + +L+ CPKLE L
Sbjct: 798 MPKLKHLEMAYHLISTSGVLQILANCPKLEFL 893
>AW203263
Length = 413
Score = 32.3 bits (72), Expect = 0.41
Identities = 31/117 (26%), Positives = 53/117 (44%), Gaps = 9/117 (7%)
Frame = +1
Query: 121 EKCSRKTLEVL-------TLASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSAC 173
EKC+ K L L L+ NPI + K + LSLS+ + + S L +C
Sbjct: 64 EKCT*KILNNLKISLTSAVLSKNPIRKIGEALMKVKSITKLSLSYCELQGIGTS--LKSC 237
Query: 174 PKLETLSIVCPEIAMSDSEASI--ELSSSSLKDFFVESYSFDKLILVADMLENLHLK 228
+L L + EI +E + +L S L + + +S K++ + L +L+L+
Sbjct: 238 VELSELRLAHNEIKSLPAELKLNSKLRSLDLGNNVITRWSELKVLELLTNLRDLNLQ 408
>TC229610 weakly similar to UP|O22512 (O22512) Grr1, partial (52%)
Length = 1428
Score = 31.6 bits (70), Expect = 0.70
Identities = 34/115 (29%), Positives = 47/115 (40%), Gaps = 7/115 (6%)
Frame = +2
Query: 130 VLTLASNPISGVEPKYHKFPCLKSLSLSFVS-ISALDLSLLLSACPKLETLSI-VCPEIA 187
V T+ +S + + H L+ L L S IS L + CP L TL+I CP I
Sbjct: 239 VSTIGDEGVSQIAKRCH---ILEKLDLCHCSSISNKGLIAIAEGCPNLTTLTIESCPNIG 409
Query: 188 MSDSEA----SIELSSSSLKDF-FVESYSFDKLILVADMLENLHLKDCSFDAFEL 237
+A +L S SLKD V + L+ A L + L+ F L
Sbjct: 410 NEGLQAIARLCTKLQSISLKDCPLVGDHGVSSLLASASNLSRVKLQTLKITDFSL 574
>BM308009 similar to PIR|D86241|D862 protein T16B5.8 [imported] -
Arabidopsis thaliana, partial (16%)
Length = 428
Score = 31.2 bits (69), Expect = 0.91
Identities = 15/50 (30%), Positives = 30/50 (60%)
Frame = +3
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDW 50
M+ +P ++ ILS + +ARDV + K+W+++ +++TL F S +
Sbjct: 30 MDSMPDAILQCILSRITNARDVSSCNCVSKRWKDS-TPYIRTLYFPRSSF 176
>TC214486
Length = 1070
Score = 30.8 bits (68), Expect = 1.2
Identities = 29/95 (30%), Positives = 44/95 (45%), Gaps = 13/95 (13%)
Frame = +1
Query: 149 PCLKSLSLSFVSISALDLSLLLSACPKLETLSI-VCPEIAMSDSEASIELSSSSLKDFF- 206
P L SL L +I+ + +S C LETL + CP+I+ S S + + SSLK F
Sbjct: 667 PRLTSLFLQSCNINEEAVEAAISKCTMLETLDVRFCPKIS-SMSMGRLRAACSSLKRIFS 843
Query: 207 -----------VESYSFDKLILVADMLENLHLKDC 230
E + + L L A+++ NL + C
Sbjct: 844 SLSAS*T*PRICEIFQWFSLGLKANLVINLVISLC 948
>TC215292 similar to UP|FENR_PEA (P10933) Ferredoxin--NADP reductase, leaf
isozyme, chloroplast precursor (FNR) , complete
Length = 1435
Score = 30.8 bits (68), Expect = 1.2
Identities = 16/49 (32%), Positives = 27/49 (54%)
Frame = +2
Query: 284 ISKASKLKKLRLWSVVFDDEDEVVDIETISVCFPRLTHLSLSYDLKDGV 332
+ K K KLRL+S+ + D +T+S+C RL + + + +L GV
Sbjct: 521 VDKNGKPHKLRLYSIASSALGDFGDSKTVSLCVKRLVYTNENGELVKGV 667
>TC215293 similar to UP|FENR_TOBAC (O04977) Ferredoxin--NADP reductase,
leaf-type isozyme, chloroplast precursor (FNR) ,
complete
Length = 1488
Score = 30.4 bits (67), Expect = 1.6
Identities = 16/49 (32%), Positives = 27/49 (54%)
Frame = +1
Query: 284 ISKASKLKKLRLWSVVFDDEDEVVDIETISVCFPRLTHLSLSYDLKDGV 332
I K K KLRL+S+ + D +T+S+C RL + + + ++ GV
Sbjct: 529 IDKNGKPHKLRLYSIASSAIGDFGDSKTVSLCVKRLVYTNENGEIVKGV 675
>TC234044
Length = 502
Score = 30.0 bits (66), Expect = 2.0
Identities = 32/130 (24%), Positives = 58/130 (44%), Gaps = 12/130 (9%)
Frame = +1
Query: 51 PLYREICSSHLEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWL--MYTRDSLRELR 108
PL + S L + ++ K + CLT + H + L MYT L E +
Sbjct: 46 PLASDAAGSELNSVEEISVPHNKIVDCLTSCIHVAHINDILEKQKGLAHMYTAFLLPEHK 225
Query: 109 YNVRTSPNFNIIEKCSR-KTLEVLTLASNPISG----VEPKYHKFP-----CLKSLSLSF 158
+ V+T+ +I E CSR + + + SN ++G V+ +H C+ ++ ++
Sbjct: 226 WTVKTTAFVSIRELCSRLQNVVKDSQGSNELAGATSFVQEIFHSLSPKILHCISTIKIAQ 405
Query: 159 VSISALDLSL 168
V +SA + L
Sbjct: 406 VHVSASECLL 435
>TC210019 similar to UP|Q84KK5 (Q84KK5) S-adenosyl-L-methionine: daidzein
7-0-methyltransferase, complete
Length = 1156
Score = 29.6 bits (65), Expect = 2.6
Identities = 16/35 (45%), Positives = 22/35 (62%)
Frame = -2
Query: 198 SSSSLKDFFVESYSFDKLILVADMLENLHLKDCSF 232
SS S+ FFV+SYSF + I+ E LHL + S+
Sbjct: 312 SSESICFFFVDSYSFKESIVFQVAHETLHLTNFSW 208
>BE659221 homologue to PIR|AB0167|AB01 ATP-dependent Clp proteinase
ATP-binding chain [imported] - Yersinia pestis (strain
CO92), partial (3%)
Length = 750
Score = 29.6 bits (65), Expect = 2.6
Identities = 25/98 (25%), Positives = 41/98 (41%), Gaps = 3/98 (3%)
Frame = -2
Query: 178 TLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSFDAFEL 237
T+ + MSD+ +S +SS + + S + A +ENL +D F +
Sbjct: 695 TVXLPANXXLMSDNSSSSPVSSXCSEFSSITSDNS-----TAASMENLVFEDDDFGFLDS 531
Query: 238 INKGTLKVLKLDDVSV---IHLDIGDNTENLEIVDVCN 272
N+ L LDD+S +D+GD + E N
Sbjct: 530 YNESFWTELNLDDISFDAPCEMDLGDTNVSFESTSCSN 417
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,589,278
Number of Sequences: 63676
Number of extensions: 342078
Number of successful extensions: 1827
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 1792
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1821
length of query: 419
length of database: 12,639,632
effective HSP length: 100
effective length of query: 319
effective length of database: 6,272,032
effective search space: 2000778208
effective search space used: 2000778208
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147007.4


