

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147007.11 - phase: 0
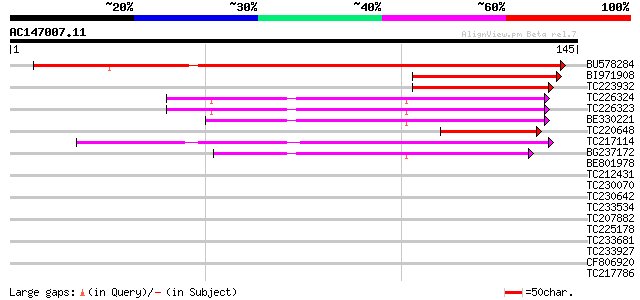
(145 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BU578284 199 4e-52
BI971908 62 7e-11
TC223932 62 1e-10
TC226324 similar to UP|Q6NM99 (Q6NM99) At1g10500, partial (71%) 60 5e-10
TC226323 similar to UP|Q6NM99 (Q6NM99) At1g10500, partial (72%) 60 5e-10
BE330221 similar to PIR|F86238|F862 protein T10O24.11 [imported]... 57 2e-09
TC220648 54 3e-08
TC217114 similar to UP|Q944M8 (Q944M8) Iron-sulfur cluster assem... 53 6e-08
BG237172 weakly similar to PIR|F86238|F86 protein T10O24.11 [imp... 42 8e-05
BE801978 weakly similar to PIR|F86238|F862 protein T10O24.11 [im... 38 0.001
TC212431 weakly similar to UP|Q9LLM1 (Q9LLM1) EF-hand Ca2+-bindi... 33 0.063
TC230070 weakly similar to GB|AAB68216.1|1244771|SCU43703 Ypl146... 31 0.24
TC230642 weakly similar to UP|O81444 (O81444) Kinase associated ... 30 0.31
TC233534 homologue to UP|Q6IID7 (Q6IID7) HDC18670, partial (16%) 28 2.0
TC207882 heat shock protein DnaJ [Glycine max] 28 2.0
TC225178 weakly similar to UP|POS5_YEAST (Q06892) POS5 protein, ... 28 2.0
TC233681 UP|Q43442 (Q43442) Phosphoinositide-specific phospholip... 28 2.0
TC233927 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis rec... 28 2.0
CF806920 27 2.6
TC217786 weakly similar to UP|Q6V9I6 (Q6V9I6) Histone deacetylas... 27 2.6
>BU578284
Length = 443
Score = 199 bits (506), Expect = 4e-52
Identities = 101/138 (73%), Positives = 120/138 (86%), Gaps = 2/138 (1%)
Frame = +3
Query: 7 RRLAPLLAARFQRNQNII--SSFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRKMKEL 64
R LA LAARF+ N N++ SS SS++S+S LHHAT SPSS EPVHIT+NCVR+MKEL
Sbjct: 3 RPLALHLAARFRHNHNLVRLSSSSSAASASTLHHATPSPSSS--EPVHITQNCVRRMKEL 176
Query: 65 DANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFNSDDRVFEKEGIKLVVDNISYDFVKG 124
+ +ESS+ G +LRLSVETGGCSGFQY FNLD+R NSDD+VFE+EG+KLVVDNISYDFVKG
Sbjct: 177 EGSESSTDGNLLRLSVETGGCSGFQYVFNLDNRINSDDKVFEREGVKLVVDNISYDFVKG 356
Query: 125 ATVDYVEELIRSAFIVSQ 142
ATVDYVEELIRSAF+V++
Sbjct: 357 ATVDYVEELIRSAFVVTE 410
>BI971908
Length = 420
Score = 62.4 bits (150), Expect = 7e-11
Identities = 30/38 (78%), Positives = 34/38 (88%)
Frame = +2
Query: 104 VFEKEGIKLVVDNISYDFVKGATVDYVEELIRSAFIVS 141
VFE++GIKLVVDNISYDFVKGATVDYVEEL +F+ S
Sbjct: 167 VFERDGIKLVVDNISYDFVKGATVDYVEELSSHSFLHS 280
Score = 34.7 bits (78), Expect(2) = 0.004
Identities = 13/20 (65%), Positives = 18/20 (90%)
Frame = +1
Query: 40 TSSPSSPDVEPVHITENCVR 59
+SS S PD+EP+H+T+NCVR
Sbjct: 106 SSSDSDPDLEPLHMTKNCVR 165
Score = 21.2 bits (43), Expect(2) = 0.004
Identities = 9/11 (81%), Positives = 10/11 (90%)
Frame = +3
Query: 28 SSSSSSSVLHH 38
SS SSSSV+HH
Sbjct: 66 SSFSSSSVVHH 98
>TC223932
Length = 530
Score = 62.0 bits (149), Expect = 1e-10
Identities = 29/36 (80%), Positives = 33/36 (91%)
Frame = +2
Query: 104 VFEKEGIKLVVDNISYDFVKGATVDYVEELIRSAFI 139
VFE++GIKLVVDNISYDFVKGATVDYVEEL +F+
Sbjct: 182 VFERDGIKLVVDNISYDFVKGATVDYVEELSSHSFL 289
Score = 34.7 bits (78), Expect(2) = 0.004
Identities = 13/20 (65%), Positives = 18/20 (90%)
Frame = +1
Query: 40 TSSPSSPDVEPVHITENCVR 59
+SS S PD+EP+H+T+NCVR
Sbjct: 121 SSSDSDPDLEPLHMTKNCVR 180
Score = 21.2 bits (43), Expect(2) = 0.004
Identities = 9/11 (81%), Positives = 10/11 (90%)
Frame = +3
Query: 28 SSSSSSSVLHH 38
SS SSSSV+HH
Sbjct: 81 SSFSSSSVVHH 113
>TC226324 similar to UP|Q6NM99 (Q6NM99) At1g10500, partial (71%)
Length = 844
Score = 59.7 bits (143), Expect = 5e-10
Identities = 32/101 (31%), Positives = 56/101 (54%), Gaps = 3/101 (2%)
Frame = +3
Query: 41 SSPSSPDVEP-VHITENCVRKMKELDANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFN 99
++P+S + P + +T+N ++ + ++ + + LR+ V+ GGCSG Y + +DR N
Sbjct: 207 AAPASGSLAPAISVTDNVLKHLNKMRSERNQD--LCLRIGVKQGGCSGMSYTMDFEDRVN 380
Query: 100 S--DDRVFEKEGIKLVVDNISYDFVKGATVDYVEELIRSAF 138
DD + E EG ++V D S F+ G +DY + LI F
Sbjct: 381 KRPDDSIIEYEGFEIVCDPKSLLFIFGMQLDYSDALIGGGF 503
>TC226323 similar to UP|Q6NM99 (Q6NM99) At1g10500, partial (72%)
Length = 810
Score = 59.7 bits (143), Expect = 5e-10
Identities = 33/101 (32%), Positives = 56/101 (54%), Gaps = 3/101 (2%)
Frame = +3
Query: 41 SSPSSPDVEP-VHITENCVRKMKELDANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFN 99
++P+S + P V +T+N ++ + ++ + + LR+ V+ GGCSG Y + +DR N
Sbjct: 246 AAPASGSLAPAVSLTDNALKHLNKMRSERNQD--LCLRIGVKQGGCSGMSYTMDFEDRAN 419
Query: 100 S--DDRVFEKEGIKLVVDNISYDFVKGATVDYVEELIRSAF 138
DD + E EG ++V D S F+ G +DY + LI F
Sbjct: 420 KRPDDSIIEYEGFEIVCDPKSLLFIFGMQLDYSDALIGGGF 542
>BE330221 similar to PIR|F86238|F862 protein T10O24.11 [imported] -
Arabidopsis thaliana, partial (79%)
Length = 295
Score = 57.4 bits (137), Expect = 2e-09
Identities = 29/90 (32%), Positives = 52/90 (57%), Gaps = 2/90 (2%)
Frame = +2
Query: 51 VHITENCVRKMKELDANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFNS--DDRVFEKE 108
V +T+N ++ + ++ + + LR+ V+ GGCSG Y +++DR N DD + E E
Sbjct: 32 VALTDNALKHLNKMRSERNQD--LCLRICVKHGGCSGMSYTMDVEDRANKRPDDAIMENE 205
Query: 109 GIKLVVDNISYDFVKGATVDYVEELIRSAF 138
G+++V D S ++ G +DY + LI +F
Sbjct: 206 GLEIVCDP*SLLYIYGVQLDYSDALIGGSF 295
>TC220648
Length = 468
Score = 53.9 bits (128), Expect = 3e-08
Identities = 26/26 (100%), Positives = 26/26 (100%)
Frame = +3
Query: 111 KLVVDNISYDFVKGATVDYVEELIRS 136
KLVVDNISYDFVKGATVDYVEELIRS
Sbjct: 3 KLVVDNISYDFVKGATVDYVEELIRS 80
>TC217114 similar to UP|Q944M8 (Q944M8) Iron-sulfur cluster assembly protein
IscA, partial (64%)
Length = 925
Score = 52.8 bits (125), Expect = 6e-08
Identities = 31/122 (25%), Positives = 61/122 (49%)
Frame = +2
Query: 18 QRNQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRKMKELDANESSSGGKMLR 77
QR+++ +SS +S + S ++ + V +T+ ++++L L+
Sbjct: 86 QRDRDSLSSMASKAMSVAAEKIATAARR---QAVSLTDAAASRIRQLLQQRQRP---FLK 247
Query: 78 LSVETGGCSGFQYAFNLDDRFNSDDRVFEKEGIKLVVDNISYDFVKGATVDYVEELIRSA 137
L V+ GC+G Y N D D + E +G+K+++D + V G +D+V++ +RS
Sbjct: 248 LGVKARGCNGLSYTLNYADEKGKFDELVEDKGVKILIDPKALMHVIGTKMDFVDDKLRSE 427
Query: 138 FI 139
FI
Sbjct: 428 FI 433
>BG237172 weakly similar to PIR|F86238|F86 protein T10O24.11 [imported] -
Arabidopsis thaliana, partial (92%)
Length = 340
Score = 42.4 bits (98), Expect = 8e-05
Identities = 24/84 (28%), Positives = 42/84 (49%), Gaps = 2/84 (2%)
Frame = +2
Query: 53 ITENCVRKMKELDANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFNS--DDRVFEKEGI 110
+T+N ++ + ++ + + +RL V GGCS Y + +DR N DD + E EG
Sbjct: 20 LTDNALKHLNKMRSERNQD--LCVRLGVRHGGCSAMSYTMDSEDRANKTPDDSIIEYEGC 193
Query: 111 KLVVDNISYDFVKGATVDYVEELI 134
++ D + G +DY + LI
Sbjct: 194 EIDSDPKILLLILGMQLDYSDALI 265
>BE801978 weakly similar to PIR|F86238|F862 protein T10O24.11 [imported] -
Arabidopsis thaliana, partial (70%)
Length = 250
Score = 38.1 bits (87), Expect = 0.001
Identities = 20/60 (33%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Frame = +2
Query: 76 LRLSVETGGCSGFQYAFNLDDRFNS--DDRVFEKEGIKLVVDNISYDFVKGATVDYVEEL 133
+R+ V+ GGCSG Y + +DR N +D + E E + V D S F+ +D+ + L
Sbjct: 56 VRIGVKQGGCSGMSYTMDFEDRVNKGPNDAIIEYECKEKVCDPNSLLFILDMQIDHSDAL 235
>TC212431 weakly similar to UP|Q9LLM1 (Q9LLM1) EF-hand Ca2+-binding protein
CCD1, partial (76%)
Length = 437
Score = 32.7 bits (73), Expect = 0.063
Identities = 19/47 (40%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Frame = -1
Query: 3 TSLFRRLAPLLAARFQRNQNIISSFS-SSSSSSVLHHATSSPSSPDV 48
+S F+ +P AA ++ N+I+ FS S+SSS+ LH + SPS P +
Sbjct: 203 SSSFKSCSPRSAALRRKLSNVITPFSLSTSSSNPLHSSFISPSPPSL 63
>TC230070 weakly similar to GB|AAB68216.1|1244771|SCU43703 Ypl146cp
{Saccharomyces cerevisiae;} , partial (5%)
Length = 1276
Score = 30.8 bits (68), Expect = 0.24
Identities = 23/59 (38%), Positives = 30/59 (49%)
Frame = -2
Query: 20 NQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRKMKELDANESSSGGKMLRL 78
N + +SS SSSSSSS ++SS S VE + C+ M +N S S GK L
Sbjct: 186 NSHSLSSSSSSSSSS----SSSSSLSSSVEIHWLRPACIGSMSVSLSNSSPSSGKFKTL 22
>TC230642 weakly similar to UP|O81444 (O81444) Kinase associated protein
phosphatase, partial (28%)
Length = 609
Score = 30.4 bits (67), Expect = 0.31
Identities = 15/35 (42%), Positives = 22/35 (62%)
Frame = -1
Query: 2 QTSLFRRLAPLLAARFQRNQNIISSFSSSSSSSVL 36
++ FR L+P + ARF + + SSFS S SS+L
Sbjct: 393 ESCCFRNLSPSILARFIPYRRVSSSFSGSPVSSIL 289
>TC233534 homologue to UP|Q6IID7 (Q6IID7) HDC18670, partial (16%)
Length = 657
Score = 27.7 bits (60), Expect = 2.0
Identities = 17/27 (62%), Positives = 20/27 (73%)
Frame = -1
Query: 20 NQNIISSFSSSSSSSVLHHATSSPSSP 46
NQ+I+SS SSSSSSS +SS SSP
Sbjct: 90 NQSIVSSSSSSSSSS-----SSSSSSP 25
>TC207882 heat shock protein DnaJ [Glycine max]
Length = 727
Score = 27.7 bits (60), Expect = 2.0
Identities = 15/63 (23%), Positives = 26/63 (40%)
Frame = +3
Query: 46 PDVEPVHITENCVRKMKELDANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFNSDDRVF 105
PDV P E ++ ++ + R + G +AFN R+N D+V
Sbjct: 285 PDVSPPGRVEEYTKRFIQVQEAYETLSDPSRRAMYDKDMAKGINFAFNARRRYNYHDQVV 464
Query: 106 EKE 108
E++
Sbjct: 465 EQK 473
>TC225178 weakly similar to UP|POS5_YEAST (Q06892) POS5 protein, partial (6%)
Length = 892
Score = 27.7 bits (60), Expect = 2.0
Identities = 19/45 (42%), Positives = 24/45 (53%)
Frame = -3
Query: 8 RLAPLLAARFQRNQNIISSFSSSSSSSVLHHATSSPSSPDVEPVH 52
R PLLAA + +S SS S +LHH + PSS D+ P H
Sbjct: 539 RPEPLLAAS---STTPLSHLHSSLPS*ILHHVPA-PSSHDLRPFH 417
>TC233681 UP|Q43442 (Q43442) Phosphoinositide-specific phospholipase C P12 ,
complete
Length = 1880
Score = 27.7 bits (60), Expect = 2.0
Identities = 12/33 (36%), Positives = 22/33 (66%)
Frame = -1
Query: 26 SFSSSSSSSVLHHATSSPSSPDVEPVHITENCV 58
+FSS++S+S + H + PSS D + ++ NC+
Sbjct: 866 NFSSTASNSGITHGDNDPSSVDSLSLSLSLNCL 768
>TC233927 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis receptor-like
kinase 2, partial (26%)
Length = 686
Score = 27.7 bits (60), Expect = 2.0
Identities = 25/77 (32%), Positives = 39/77 (50%), Gaps = 6/77 (7%)
Frame = +1
Query: 72 GGKMLRLSVETGGCSGFQYAFNLDDRFNSDDRVFEKEGIKLVVDNISYDFVKGAT----- 126
G M+ L + TG Q AF+L RF D+ + E +K++V + + + A
Sbjct: 175 GYGMMLLEIITG-----QRAFDLA-RFARDEDIMLLEWVKVLVKDKKLETLVDANLRGNC 336
Query: 127 -VDYVEELIRSAFIVSQ 142
++ VEELIR A I +Q
Sbjct: 337 DIEEVEELIRVALICTQ 387
>CF806920
Length = 710
Score = 27.3 bits (59), Expect = 2.6
Identities = 17/48 (35%), Positives = 24/48 (49%), Gaps = 5/48 (10%)
Frame = +2
Query: 28 SSSSSSSVLHHATSSPSSP-----DVEPVHITENCVRKMKELDANESS 70
S S S V HH TSS SSP + + + +R++K+ NE S
Sbjct: 77 SCGSGSGVQHHTTSSSSSPAQSRLQRQKLEVYNEILRRLKD-SGNEKS 217
>TC217786 weakly similar to UP|Q6V9I6 (Q6V9I6) Histone deacetylase, partial
(48%)
Length = 1193
Score = 27.3 bits (59), Expect = 2.6
Identities = 16/46 (34%), Positives = 25/46 (53%)
Frame = -3
Query: 26 SFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRKMKELDANESSS 71
S S SSS S +SSPS + P ++ + + + D++ESSS
Sbjct: 630 SSSDSSSPSSSSSLSSSPSLLSMSPFAVS*SSLESSSDSDSDESSS 493
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.131 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,689,148
Number of Sequences: 63676
Number of extensions: 83584
Number of successful extensions: 1234
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 956
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1119
length of query: 145
length of database: 12,639,632
effective HSP length: 88
effective length of query: 57
effective length of database: 7,036,144
effective search space: 401060208
effective search space used: 401060208
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC147007.11


