

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147002.5 + phase: 0
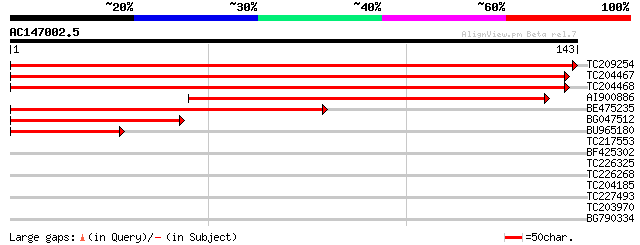
(143 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209254 homologue to UP|Q9FUN3 (Q9FUN3) QM-like protein, partia... 271 1e-73
TC204467 homologue to UP|RL10_VITRI (Q9SPB3) 60S ribosomal prote... 264 1e-71
TC204468 homologue to UP|RL10_SOLME (P93847) 60S ribosomal prote... 262 4e-71
AI900886 similar to SP|Q40592|RL10_ 60S ribosomal protein L10 (Q... 125 9e-30
BE475235 similar to SP|Q9M5M7|RL10 60S ribosomal protein L10. [L... 72 7e-14
BG047512 weakly similar to SP|Q9M5M7|RL10 60S ribosomal protein ... 45 9e-06
BU965180 similar to SP|Q9SPB3|RL10_ 60S ribosomal protein L10 (Q... 45 1e-05
TC217553 28 1.9
BF425302 homologue to GP|28805416|dbj protease DO {Vibrio paraha... 26 5.7
TC226325 UP|Q6SQJ1 (Q6SQJ1) NBS-LRR type disease resistance prot... 26 7.4
TC226268 similar to UP|Q9SXL2 (Q9SXL2) Serine decarboxylase, par... 26 7.4
TC204185 homologue to UP|TBB1_SOYBN (P12459) Tubulin beta-1 chai... 26 7.4
TC227493 similar to UP|FABG_CUPLA (P28643) 3-oxoacyl-[acyl-carri... 26 7.4
TC203970 homologue to UP|Q6UCJ3 (Q6UCJ3) Ubiquitin extension pro... 26 7.4
BG790334 weakly similar to GP|2160154|gb| F19K23.18 gene product... 25 9.7
>TC209254 homologue to UP|Q9FUN3 (Q9FUN3) QM-like protein, partial (96%)
Length = 1060
Score = 271 bits (692), Expect = 1e-73
Identities = 130/143 (90%), Positives = 135/143 (93%)
Frame = +2
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+KFAGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GTCARV+IGQVL
Sbjct: 374 MAKFAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVNIGQVL 553
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HAQEALRRAKFKFPGRQKII+SRKWGFTK S +EYLKLKSENRIVPDGV
Sbjct: 554 LSVRCKDANSHHAQEALRRAKFKFPGRQKIILSRKWGFTKFSRSEYLKLKSENRIVPDGV 733
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAKVLGCHGPLANR+PGRAFLPA
Sbjct: 734 NAKVLGCHGPLANREPGRAFLPA 802
>TC204467 homologue to UP|RL10_VITRI (Q9SPB3) 60S ribosomal protein L10 (QM
protein homolog), partial (98%)
Length = 914
Score = 264 bits (674), Expect = 1e-71
Identities = 126/141 (89%), Positives = 132/141 (93%)
Frame = +2
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+KFAGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GTCARV+IGQVL
Sbjct: 278 MAKFAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVAIGQVL 457
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HAQEALRRAKFKFPGRQKIIVSRKWGFTK S ++YLK KSENRIVPDGV
Sbjct: 458 LSVRCKDSNSHHAQEALRRAKFKFPGRQKIIVSRKWGFTKFSRSDYLKFKSENRIVPDGV 637
Query: 121 NAKVLGCHGPLANRQPGRAFL 141
NAK+LGCHGPLANR+PGRAFL
Sbjct: 638 NAKLLGCHGPLANREPGRAFL 700
>TC204468 homologue to UP|RL10_SOLME (P93847) 60S ribosomal protein L10
(EQM), complete
Length = 1127
Score = 262 bits (670), Expect = 4e-71
Identities = 126/141 (89%), Positives = 132/141 (93%)
Frame = +1
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+KFAGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GTCARV+IGQVL
Sbjct: 313 MAKFAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGTCARVAIGQVL 492
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HAQEALRRAKFKFPGRQKIIVSRKWGFTK S A+YLK KSE+RIVPDGV
Sbjct: 493 LSVRCKDSNSHHAQEALRRAKFKFPGRQKIIVSRKWGFTKFSRADYLKYKSESRIVPDGV 672
Query: 121 NAKVLGCHGPLANRQPGRAFL 141
NAK+LGCHGPLANRQPG+AFL
Sbjct: 673 NAKLLGCHGPLANRQPGQAFL 735
>AI900886 similar to SP|Q40592|RL10_ 60S ribosomal protein L10 (QM protein
homolog) (Fragment). [Common tobacco] {Nicotiana
tabacum}, partial (55%)
Length = 274
Score = 125 bits (313), Expect = 9e-30
Identities = 63/91 (69%), Positives = 68/91 (74%)
Frame = +2
Query: 46 KPLGTCARVSIGQVLLSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAE 105
KP GTCARV+IGQVLLSVRCK N HAQEALRRAKFKF RQKII SR GFTKLS ++
Sbjct: 2 KPHGTCARVAIGQVLLSVRCKDSNQHHAQEALRRAKFKFARRQKIIASRTCGFTKLSRSD 181
Query: 106 YLKLKSENRIVPDGVNAKVLGCHGPLANRQP 136
YLK ENR V DG NA + GCH P R+P
Sbjct: 182 YLKF*VENRCVRDGANAMLHGCHAPFGYREP 274
>BE475235 similar to SP|Q9M5M7|RL10 60S ribosomal protein L10. [Leafy spurge]
{Euphorbia esula}, partial (70%)
Length = 463
Score = 72.4 bits (176), Expect = 7e-14
Identities = 43/80 (53%), Positives = 51/80 (63%)
Frame = +2
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M+K AGKD L V +HPFH LRIN M AGA+R+ T +RGA P GT AR +IGQV
Sbjct: 224 MAKSAGKDASLLTVTLHPFHALRINTMPPRAGANRIPTVLRGAS*PPYGTWARYAIGQVT 403
Query: 61 LSVRCKSGNGDHAQEALRRA 80
SVR K + A +AL RA
Sbjct: 404 PSVR*KDRSSHLAWDALWRA 463
>BG047512 weakly similar to SP|Q9M5M7|RL10 60S ribosomal protein L10. [Leafy
spurge] {Euphorbia esula}, partial (50%)
Length = 376
Score = 45.4 bits (106), Expect = 9e-06
Identities = 24/44 (54%), Positives = 30/44 (67%)
Frame = +2
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAF 44
M AGK + VRVHP++VL IN MLS + A+ LQT M+GAF
Sbjct: 203 MD*LAGKGCLNYIVRVHPYNVLMIN*MLSYSSANMLQTVMKGAF 334
>BU965180 similar to SP|Q9SPB3|RL10_ 60S ribosomal protein L10 (QM protein
homolog). [Frost grape Vitis vulpina] {Vitis riparia},
partial (45%)
Length = 430
Score = 45.1 bits (105), Expect = 1e-05
Identities = 21/29 (72%), Positives = 24/29 (82%)
Frame = +3
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLS 29
+S A + FHLRVRVHPFHVLRINKML+
Sbjct: 288 VSSEALEGAFHLRVRVHPFHVLRINKMLA 374
Score = 35.8 bits (81), Expect = 0.007
Identities = 15/18 (83%), Positives = 16/18 (88%)
Frame = +1
Query: 124 VLGCHGPLANRQPGRAFL 141
+LGCHGPLAN QPGRA L
Sbjct: 373 LLGCHGPLANCQPGRACL 426
>TC217553
Length = 1150
Score = 27.7 bits (60), Expect = 1.9
Identities = 14/34 (41%), Positives = 18/34 (52%)
Frame = +2
Query: 46 KPLGTCARVSIGQVLLSVRCKSGNGDHAQEALRR 79
K G +++ VLL +R G GDH E LRR
Sbjct: 215 KASGRAPPLAVAGVLLRLRGGGGAGDHGGEGLRR 316
>BF425302 homologue to GP|28805416|dbj protease DO {Vibrio parahaemolyticus},
partial (2%)
Length = 269
Score = 26.2 bits (56), Expect = 5.7
Identities = 12/26 (46%), Positives = 17/26 (65%)
Frame = +3
Query: 20 HVLRINKMLSCAGADRLQTGMRGAFG 45
H LR+N+ LS + +DR +TG G G
Sbjct: 132 HKLRVNRPLSLSLSDRSETGGSGVGG 209
>TC226325 UP|Q6SQJ1 (Q6SQJ1) NBS-LRR type disease resistance protein RPG1-B,
complete
Length = 3754
Score = 25.8 bits (55), Expect = 7.4
Identities = 19/55 (34%), Positives = 26/55 (46%), Gaps = 4/55 (7%)
Frame = +3
Query: 6 GKDTFHLRVRVHPFHVLRINKMLSC----AGADRLQTGMRGAFGKPLGTCARVSI 56
G + + RV +PFH+ +SC AG + +TG R L T AR SI
Sbjct: 3537 GSNAYQRRVCPNPFHLCGFGGTVSCSKNVAGNPKAKTGQR------LLTFARCSI 3683
>TC226268 similar to UP|Q9SXL2 (Q9SXL2) Serine decarboxylase, partial (81%)
Length = 1703
Score = 25.8 bits (55), Expect = 7.4
Identities = 12/39 (30%), Positives = 20/39 (50%)
Frame = -3
Query: 77 LRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRI 115
++ FK PG I+ KW +K+ + + LKL+ I
Sbjct: 549 IQNTNFKLPGVDTIVAFNKW-ISKVVYGKVLKLRERTII 436
>TC204185 homologue to UP|TBB1_SOYBN (P12459) Tubulin beta-1 chain, complete
Length = 1732
Score = 25.8 bits (55), Expect = 7.4
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 12/49 (24%)
Frame = -2
Query: 66 KSGNGDHAQEALRRAKFKFPGRQK------IIVSRKW------GFTKLS 102
+ G G +LRR++F PG+++ I+V KW GF LS
Sbjct: 936 RRGKGMRFTASLRRSEFSCPGKRRQHVTPLIVVEIKWFRSPKLGFVSLS 790
>TC227493 similar to UP|FABG_CUPLA (P28643) 3-oxoacyl-[acyl-carrier-protein]
reductase, chloroplast precursor (3-ketoacyl-acyl
carrier protein reductase) , partial (83%)
Length = 1452
Score = 25.8 bits (55), Expect = 7.4
Identities = 18/51 (35%), Positives = 23/51 (44%), Gaps = 2/51 (3%)
Frame = +1
Query: 76 ALRRAKFKFPGRQKIIVSRKWG--FTKLSHAEYLKLKSENRIVPDGVNAKV 124
ALR A F G +KI R+W T L L+ +S GV A+V
Sbjct: 208 ALRTANFGASGNRKIGQIRQWSPILTNLRPVSGLRHRSNTPFSSSGVRAQV 360
>TC203970 homologue to UP|Q6UCJ3 (Q6UCJ3) Ubiquitin extension protein,
partial (98%)
Length = 748
Score = 25.8 bits (55), Expect = 7.4
Identities = 15/40 (37%), Positives = 19/40 (47%)
Frame = -1
Query: 29 SCAGADRLQTGMRGAFGKPLGTCARVSIGQVLLSVRCKSG 68
SCA R G RG P G C+R G+ + R +SG
Sbjct: 307 SCASWRRRGGGGRGEAWTPSGCCSRRGCGRPPAAYRRRSG 188
>BG790334 weakly similar to GP|2160154|gb| F19K23.18 gene product
{Arabidopsis thaliana}, partial (19%)
Length = 390
Score = 25.4 bits (54), Expect = 9.7
Identities = 17/73 (23%), Positives = 33/73 (44%), Gaps = 2/73 (2%)
Frame = +2
Query: 13 RVRVHPFHVLRINKMLSCAGADRLQTGMR--GAFGKPLGTCARVSIGQVLLSVRCKSGNG 70
R+++HP ++ I+ + +CA A ++ G R + G RV L+ + + G
Sbjct: 11 RLKIHPTYITFISVLNACAHAGLVEEGWRQFKSMINDYGIEPRVEHFASLVDILGRQGQL 190
Query: 71 DHAQEALRRAKFK 83
A + + FK
Sbjct: 191QEAMDLINTMPFK 229
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.140 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,428,812
Number of Sequences: 63676
Number of extensions: 79986
Number of successful extensions: 454
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 453
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 454
length of query: 143
length of database: 12,639,632
effective HSP length: 88
effective length of query: 55
effective length of database: 7,036,144
effective search space: 386987920
effective search space used: 386987920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC147002.5


