

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.3 + phase: 0 /pseudo
(651 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
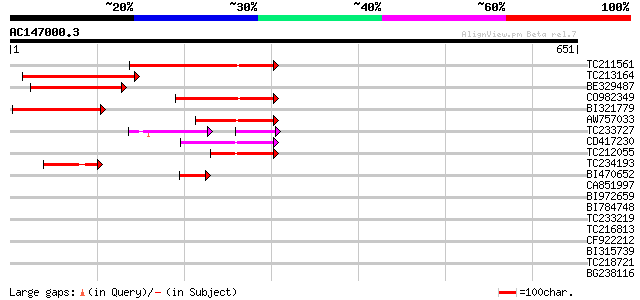
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 167 1e-41
TC213164 140 1e-33
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 123 2e-28
CO982349 107 2e-23
BI321779 101 9e-22
AW757033 87 3e-17
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 62 2e-12
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 65 7e-11
TC212055 62 1e-09
TC234193 52 8e-07
BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase ... 47 2e-05
CA851997 37 0.036
BI972659 36 0.061
BI784748 36 0.061
TC233219 weakly similar to UP|Q8LKI9 (Q8LKI9) NBS/LRR resistance... 34 0.18
TC216813 similar to UP|WR11_ARATH (Q9SV15) Probable WRKY transcr... 32 0.89
CF922212 32 1.2
BI315739 26 2.0
TC218721 similar to UP|Q9FJ27 (Q9FJ27) Polygalacturonase-like pr... 30 4.4
BG238116 weakly similar to SP|Q9M7Q7|PCN1_ Proliferating cellula... 30 4.4
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 167 bits (424), Expect = 1e-41
Identities = 86/171 (50%), Positives = 117/171 (68%)
Frame = +1
Query: 138 LDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQKWISEC 197
L LIANEV+D+A+R +K L+F+VD+EKAYDSV +L +M M+F W KWI EC
Sbjct: 1 LHSALIANEVIDEAKRSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEEC 180
Query: 198 IGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFNGFLV 257
+ +A +VLVNG P EF +RGLRQGDPL+PFLF + AEG N LM++ L+ +LV
Sbjct: 181 VKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYLV 360
Query: 258 GRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKS 308
G G V ++ LQ+ADDT+ GE + NV +++ +L FE VS L++NF KS
Sbjct: 361 GANG-VPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKS 510
>TC213164
Length = 446
Score = 140 bits (354), Expect = 1e-33
Identities = 60/135 (44%), Positives = 99/135 (72%)
Frame = +3
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L F +EV+ +WD + KSPGP+G++F FIK FWD++K D ++FL +F+ NG +K
Sbjct: 18 LVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLKR 197
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
N+ F+ALIPKV++P LN+Y PISL+GC+YK++AK+L+ R + V+ +++ + Q+ F++G
Sbjct: 198 SNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFMEG 377
Query: 135 N*ILDGILIANEVVD 149
L ++IANE+++
Sbjct: 378 RHTLHNVVIANEIME 422
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 123 bits (309), Expect = 2e-28
Identities = 59/114 (51%), Positives = 80/114 (69%), Gaps = 3/114 (2%)
Frame = -2
Query: 24 VKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKGVNSTFIALI 83
+K AVW C S KSPGP+G++F FIK FW+ +K D ++FL +FH NG KG N++FIALI
Sbjct: 375 IKSAVWQCGSVKSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALI 196
Query: 84 PKVNNPQRLNDYMPISLVGCLYKVLAKVLA-NRLRNVIGSVVSD--SQSAFIKG 134
PKV +PQ LND+ PISL+GC+YK++AK+ N + +V D + AF KG
Sbjct: 195 PKVKHPQALNDFRPISLIGCVYKIVAKIXXQNXIEKXCCRMVID*SDKLAFXKG 34
>CO982349
Length = 795
Score = 107 bits (266), Expect = 2e-23
Identities = 54/118 (45%), Positives = 78/118 (65%)
Frame = +3
Query: 191 QKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQ 250
Q WI C+ +A ++LVN P+ EFS +RGLRQGDPL+P LF + AEG LM++ +
Sbjct: 63 QHWIRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRK 242
Query: 251 LFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKS 308
FN FLVG+ E ++ LQ+ADDT+ E + N++ ++ +L FE SGLK+NF +S
Sbjct: 243 RFNSFLVGKNKE-PVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAES 413
>BI321779
Length = 421
Score = 101 bits (252), Expect = 9e-22
Identities = 48/107 (44%), Positives = 69/107 (63%)
Frame = +2
Query: 4 FRKLSLREAGNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLV 63
F S + L + F EE+K+ + DC+S KSPGP+G S IK FW+++K++ + F
Sbjct: 50 FLSFSNEDNAMLIQDFEVEEIKKVI*DCESSKSPGPDGYSLLLIKIFWEVIKEEVINFFK 229
Query: 64 DFHRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAK 110
+FH+ L +G N +FIALI K +NP + Y PISLVGCLYK++ K
Sbjct: 230 EFHKFDFLPRGTNRSFIALIAKCDNPLDIGQYCPISLVGCLYKIV*K 370
>AW757033
Length = 441
Score = 86.7 bits (213), Expect = 3e-17
Identities = 46/95 (48%), Positives = 62/95 (64%)
Frame = -2
Query: 214 EFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFNGFLVGRVGEVNLTHLQFADD 273
EFS RGLRQGDPL+P LF +AAEG LM++ + F VG+ + ++ LQ+ADD
Sbjct: 434 EFSPHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKY-KKPVSILQYADD 258
Query: 274 TLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKS 308
T+ GE + NVR ++ +L FE SGLK+NF KS
Sbjct: 257 TIFFGEATMENVRVIKTILRGFELASGLKINFAKS 153
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 62.0 bits (149), Expect(2) = 2e-12
Identities = 35/100 (35%), Positives = 59/100 (59%), Gaps = 4/100 (4%)
Frame = -3
Query: 137 ILDGILIANEVVDDARRMDKEL----LLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQK 192
I D + +EV++ +DK++ L ++D KA+D++D +L V+ + +
Sbjct: 834 IKDCTCVTSEVIN---MLDKKVFGGNLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCN 664
Query: 193 WISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLF 232
WI + +AK ++ VNG P+ FS +RG+RQGDPLSP L+
Sbjct: 663 WIRVILLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLY 544
Score = 28.5 bits (62), Expect(2) = 2e-12
Identities = 13/51 (25%), Positives = 28/51 (54%)
Frame = -2
Query: 260 VGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSML 310
+G + +H+ ADD +I + + NV ++ L+++ G ++ HKS +
Sbjct: 469 MGSLTRSHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKI 317
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 65.5 bits (158), Expect = 7e-11
Identities = 38/112 (33%), Positives = 61/112 (53%)
Frame = -3
Query: 197 CIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFNGFL 256
C+ T ++L NG ++ FS G+RQ DP++P+LF+L E + L+ + +L
Sbjct: 668 CMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKKL*RSIQ 489
Query: 257 VGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKS 308
V R G + ++HL F DD ++ E + V + L LF + SG KVN K+
Sbjct: 488 VNR-GGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKT 336
>TC212055
Length = 776
Score = 61.6 bits (148), Expect = 1e-09
Identities = 30/78 (38%), Positives = 52/78 (66%)
Frame = +1
Query: 231 LFLLAAEGFNVLMKQMAGAQLFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRA 290
LF + AEG LM++ L++ +VG+ ++ + LQ+ADDT+ +GE + NV ++++
Sbjct: 1 LFNIVAEGLTGLMREALDKSLYSSLMVGK-NKIPVNILQYADDTIFLGEATMQNVMTIKS 177
Query: 291 VLMLFEEVSGLKVNFHKS 308
+L +FE SGLK++F KS
Sbjct: 178 MLRVFELASGLKIHFAKS 231
>TC234193
Length = 697
Score = 52.0 bits (123), Expect = 8e-07
Identities = 25/67 (37%), Positives = 44/67 (65%)
Frame = -3
Query: 40 NGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPIS 99
NG +FNFIK+ +++K+D ++ + +FH +G L +G N++F+ + P L D+ P++
Sbjct: 194 NGYNFNFIKKC*EVMKEDVVRAIQEFHSHGCLSRGTNASFL-----THPP*GLGDFRPVT 30
Query: 100 LVGCLYK 106
LVG L K
Sbjct: 29 LVGSLCK 9
>BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase (EC
2.7.7.49) (clone AmLi2) - garden snapdragon (fragment),
partial (22%)
Length = 111
Score = 47.4 bits (111), Expect = 2e-05
Identities = 20/35 (57%), Positives = 24/35 (68%)
Frame = +1
Query: 196 ECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPF 230
EC+ +A ++LVNG P EEF RGLRQG P PF
Sbjct: 4 ECLSSASISILVNGSPTEEFKPXRGLRQGGPFGPF 108
>CA851997
Length = 636
Score = 36.6 bits (83), Expect = 0.036
Identities = 26/84 (30%), Positives = 40/84 (46%), Gaps = 2/84 (2%)
Frame = +1
Query: 39 PNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPI 98
P+G + F RFW + +D Q + G VN T IA+I ++ R
Sbjct: 316 PDGFNLGFYHRFWGMCGEDVFQACCMWLAEGAFPSSVNDTTIAIILEI**S*RYERS*TN 495
Query: 99 SLVGCLYKV--LAKVLANRLRNVI 120
L +K+ L++VLA RL+NV+
Sbjct: 496 LLCNGGFKILFLSEVLAKRLKNVL 567
>BI972659
Length = 453
Score = 35.8 bits (81), Expect = 0.061
Identities = 15/32 (46%), Positives = 24/32 (74%)
Frame = +1
Query: 277 IGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKS 308
+GE SW N+ ++++L FE VSGL++N+ KS
Sbjct: 1 VGEASWDNIIVLKSMLRGFEMVSGLRINYAKS 96
>BI784748
Length = 259
Score = 35.8 bits (81), Expect = 0.061
Identities = 16/33 (48%), Positives = 25/33 (75%)
Frame = +2
Query: 128 QSAFIKGN*ILDGILIANEVVDDARRMDKELLL 160
QSAFI+G +L ++IANEV+D+A+R K ++
Sbjct: 14 QSAFIQGRHMLHSMMIANEVIDEAKRGQKPSIM 112
>TC233219 weakly similar to UP|Q8LKI9 (Q8LKI9) NBS/LRR resistance
protein-like protein (Fragment), partial (74%)
Length = 781
Score = 34.3 bits (77), Expect = 0.18
Identities = 23/86 (26%), Positives = 36/86 (41%)
Frame = +3
Query: 194 ISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFN 253
I + +V G ++F I GL QG LSP+LF L +
Sbjct: 54 IQDMYDRVSTSVRTQGGESDDFPITIGLHQGSTLSPYLFTLILD---------------- 185
Query: 254 GFLVGRVGEVNLTHLQFADDTLIIGE 279
L ++ E+ + FADD +++GE
Sbjct: 186 -VLTEQIQEIVSRCMLFADDIVLLGE 260
>TC216813 similar to UP|WR11_ARATH (Q9SV15) Probable WRKY transcription
factor 11 (WRKY DNA-binding protein 11), partial (46%)
Length = 1173
Score = 32.0 bits (71), Expect = 0.89
Identities = 18/39 (46%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Frame = -2
Query: 551 SWRWRRRVRWRRCGGWGGAVMEGSG-FGDVVCLHGRRRV 588
SW+ RWRRCG GG ++G G G V C G RR+
Sbjct: 266 SWK-----RWRRCGECGGGKVDGRGDGGGVPCP*GLRRL 165
>CF922212
Length = 445
Score = 31.6 bits (70), Expect = 1.2
Identities = 21/48 (43%), Positives = 24/48 (49%)
Frame = -3
Query: 427 LGLIGIRFVYQRKKGGWGLGG*ELLIWHC*ENGVGRC**IKRGCGIEC 474
LG GI+ YQR+K GWG G * LI E G G K G+ C
Sbjct: 359 LG*TGIQCAYQRRKRGWGTGI*GNLIMLFLEKGDGIYSITKGNWGLGC 216
>BI315739
Length = 442
Score = 25.8 bits (55), Expect(2) = 2.0
Identities = 15/44 (34%), Positives = 23/44 (52%)
Frame = +2
Query: 265 LTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKS 308
++HL F D+ L+ + + V+ +R VL F GLK KS
Sbjct: 164 ISHLFFPDNCLLFVKATSSQVKLVRDVLQQFCLT*GLKAGLQKS 295
Score = 23.5 bits (49), Expect(2) = 2.0
Identities = 10/36 (27%), Positives = 18/36 (49%)
Frame = +3
Query: 226 PLSPFLFLLAAEGFNVLMKQMAGAQLFNGFLVGRVG 261
PLSP+LF + E +L++ + + + R G
Sbjct: 48 PLSPYLF*ICMEKLAILIQSKVNEETWEPIKISRNG 155
>TC218721 similar to UP|Q9FJ27 (Q9FJ27) Polygalacturonase-like protein,
partial (53%)
Length = 1128
Score = 29.6 bits (65), Expect = 4.4
Identities = 11/25 (44%), Positives = 16/25 (64%)
Frame = -3
Query: 338 LTYWRGFSKIEFLETCCRPYCGTFV 362
++YW+ K EFL C +P C TF+
Sbjct: 793 ISYWQISVKGEFLNPCIKPACKTFI 719
>BG238116 weakly similar to SP|Q9M7Q7|PCN1_ Proliferating cellular nuclear
antigen 1 (PCNA 1). [Mouse-ear cress] {Arabidopsis
thaliana}, partial (22%)
Length = 544
Score = 29.6 bits (65), Expect = 4.4
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Frame = +2
Query: 69 GKLIKGVNSTFIALIPKVNNPQRLNDYM--PISLVGCLYKVLAKVLANRLRN 118
G +K +NST AL+P + P+ N Y PI+ G A++L + +N
Sbjct: 98 GFSLKALNSTLCALVPLLLRPEGFNPYRWNPITSWGLTLNTWARILRSPGKN 253
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.344 0.156 0.540
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,087,586
Number of Sequences: 63676
Number of extensions: 552652
Number of successful extensions: 7165
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 5187
Number of HSP's successfully gapped in prelim test: 197
Number of HSP's that attempted gapping in prelim test: 1486
Number of HSP's gapped (non-prelim): 5885
length of query: 651
length of database: 12,639,632
effective HSP length: 103
effective length of query: 548
effective length of database: 6,081,004
effective search space: 3332390192
effective search space used: 3332390192
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147000.3


