

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146940.1 - phase: 0
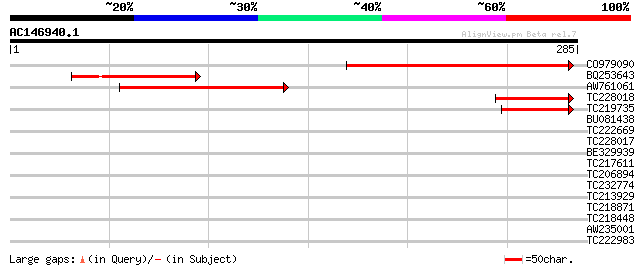
(285 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CO979090 187 6e-48
BQ253643 67 7e-12
AW761061 66 2e-11
TC228018 55 5e-08
TC219735 44 1e-04
BU081438 similar to GP|6520167|dbj ZW18 {Arabidopsis thaliana}, ... 37 0.010
TC222669 36 0.017
TC228017 35 0.051
BE329939 similar to GP|6520167|dbj| ZW18 {Arabidopsis thaliana},... 34 0.066
TC217611 34 0.086
TC206894 similar to GB|AAQ22603.1|33589674|BT010134 At2g02560 {A... 31 0.56
TC232774 30 0.96
TC213929 weakly similar to UP|Q94G63 (Q94G63) Ethylene-induced e... 30 0.96
TC218871 30 0.96
TC218448 weakly similar to UP|DR29_ARATH (Q9SZA7) Probable disea... 30 1.2
AW235001 28 4.7
TC222983 28 6.2
>CO979090
Length = 686
Score = 187 bits (474), Expect = 6e-48
Identities = 89/114 (78%), Positives = 99/114 (86%)
Frame = -3
Query: 170 QISNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFL 229
QIS+QECH RKYEGKIAGLEPINFI ATPHLG RGHKQVP+ CGF+SLEK SR++
Sbjct: 684 QISDQECHDRKYEGKIAGLEPINFIXXATPHLGSRGHKQVPMFCGFYSLEKAVSRVAGVF 505
Query: 230 GKTGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
GKTGKHLFLTD N KPPLLLQMV DSEDIKF+SALRSFKRRVAYAN+ YD+++
Sbjct: 504 GKTGKHLFLTDRDNGKPPLLLQMVHDSEDIKFLSALRSFKRRVAYANVLYDQLV 343
>BQ253643
Length = 344
Score = 67.4 bits (163), Expect = 7e-12
Identities = 32/65 (49%), Positives = 44/65 (67%)
Frame = +1
Query: 32 GCFRIQHDATGDGFDIEVVDASGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDV 91
GCF+ + D+ G F + A+ P HL+IMVNG+IGSA +W+YAA+QF+K+ P V
Sbjct: 52 GCFKAEADSAGQDF-FDAAAAAAPNPAPHHLVIMVNGIIGSAADWRYAAEQFVKKLPDKV 228
Query: 92 IVHCS 96
IVH S
Sbjct: 229 IVHRS 243
>AW761061
Length = 316
Score = 65.9 bits (159), Expect = 2e-11
Identities = 29/85 (34%), Positives = 54/85 (63%)
Frame = +2
Query: 56 RSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRL 115
+++P HL+++V+G++ S +W A + +R + I++ S N+ T T +D G RL
Sbjct: 62 KNDPDHLLVLVHGILASTADWTDAEAELKRRLGKNFIIYVSSSNTYTKTLTAIDGAGKRL 241
Query: 116 AEEVISVIKRHPSVRKISFIAHSLG 140
A+E ++K+ S+++I F+AHSLG
Sbjct: 242 ADER*LIVKKTKSLKRIFFLAHSLG 316
>TC228018
Length = 760
Score = 54.7 bits (130), Expect = 5e-08
Identities = 25/39 (64%), Positives = 32/39 (81%)
Frame = +1
Query: 245 KPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
KPPLL +MV+D D+ FMSALR+FKRR AY+N+ YD I+
Sbjct: 7 KPPLLKRMVQDYGDLYFMSALRAFKRRFAYSNVDYDHIV 123
>TC219735
Length = 628
Score = 43.5 bits (101), Expect = 1e-04
Identities = 19/36 (52%), Positives = 28/36 (77%)
Frame = +1
Query: 248 LLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
LLL+M DS+D KF+SAL +F+ R+ YAN+ YD ++
Sbjct: 1 LLLRMASDSDDGKFLSALGAFRCRIIYANVSYDHMV 108
>BU081438 similar to GP|6520167|dbj ZW18 {Arabidopsis thaliana}, partial
(14%)
Length = 425
Score = 37.0 bits (84), Expect = 0.010
Identities = 29/98 (29%), Positives = 50/98 (50%), Gaps = 9/98 (9%)
Frame = +2
Query: 62 LIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVIS 121
+++ V+G G + + Q+L P V SE N + D ++ G+RLA+EVIS
Sbjct: 107 IVVFVHGFQGHHLDLRLIRNQWLLIDP-KVEFLMSETNEDKTSGDFREM-GHRLAQEVIS 280
Query: 122 VIKRHPSVR---------KISFIAHSLGGLIARYAIAK 150
+++ ++SF+ HS+G LI R A+A+
Sbjct: 281 FVRKKMDKASRYGNLGDIRLSFVGHSIGNLIIRTALAE 394
>TC222669
Length = 653
Score = 36.2 bits (82), Expect = 0.017
Identities = 26/63 (41%), Positives = 36/63 (56%), Gaps = 9/63 (14%)
Frame = +2
Query: 96 SECNSSTLTFDGVDVTGNRLAEEVISVIK-------RHPSVR--KISFIAHSLGGLIARY 146
SE N TF G+RLA+EVI+ +K R+ S+ ++SF+ HS+G LI R
Sbjct: 65 SEANEDK-TFGDFREMGHRLAKEVIAFLKSKMDKASRYGSLGDIRLSFVGHSIGNLIIRT 241
Query: 147 AIA 149
AIA
Sbjct: 242 AIA 250
>TC228017
Length = 752
Score = 34.7 bits (78), Expect = 0.051
Identities = 15/23 (65%), Positives = 19/23 (82%)
Frame = +3
Query: 261 FMSALRSFKRRVAYANIRYDRIL 283
FMSAL +FKRR AY+N+ YD I+
Sbjct: 9 FMSALCAFKRRFAYSNVDYDHIV 77
>BE329939 similar to GP|6520167|dbj| ZW18 {Arabidopsis thaliana}, partial
(12%)
Length = 386
Score = 34.3 bits (77), Expect = 0.066
Identities = 22/63 (34%), Positives = 35/63 (54%), Gaps = 9/63 (14%)
Frame = +1
Query: 96 SECNSSTLTFDGVDVTGNRLAEEVISVIKRHPSVR---------KISFIAHSLGGLIARY 146
SE N + D ++ G+RLA+EVIS +++ ++SF+ HS+G LI R
Sbjct: 40 SETNEDKTSGDFREM-GHRLAQEVISFVRKKMDKASRYGNLGDIRLSFVGHSIGNLIIRT 216
Query: 147 AIA 149
A+A
Sbjct: 217 ALA 225
>TC217611
Length = 639
Score = 33.9 bits (76), Expect = 0.086
Identities = 28/98 (28%), Positives = 52/98 (52%), Gaps = 9/98 (9%)
Frame = +3
Query: 62 LIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVIS 121
+++ V+G G+ + + Q+L P + S+ N + D ++ G RLA+EVI
Sbjct: 141 IVVFVHGFQGNHLDLRLVRNQWLLIDP-KIQFLMSQANEDKTSGDFREM-GFRLAQEVIL 314
Query: 122 VIK-------RHPSVR--KISFIAHSLGGLIARYAIAK 150
+K R+ +++ K+SF+ HS+G LI R A+ +
Sbjct: 315 FLKKKMDKASRNGNLKDIKLSFVGHSIGNLIIRTALTE 428
>TC206894 similar to GB|AAQ22603.1|33589674|BT010134 At2g02560 {Arabidopsis
thaliana;} , partial (44%)
Length = 1951
Score = 31.2 bits (69), Expect = 0.56
Identities = 21/67 (31%), Positives = 33/67 (48%), Gaps = 2/67 (2%)
Frame = +1
Query: 130 RKISFIAHSLGGLIARYAI--AKLYERDISKELSQGNVHCEGQISNQECHVRKYEGKIAG 187
+K + HSL +I R ++ A+ E + K L+ HCE + V + GKIA
Sbjct: 700 KKQYLLLHSLKEVIVRQSVDKAEFQESSVEKILNLLFNHCESEEEGVRNVVAECLGKIAL 879
Query: 188 LEPINFI 194
+EP+ I
Sbjct: 880 IEPVKLI 900
>TC232774
Length = 589
Score = 30.4 bits (67), Expect = 0.96
Identities = 25/97 (25%), Positives = 47/97 (47%), Gaps = 1/97 (1%)
Frame = +2
Query: 59 PTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDV-IVHCSECNSSTLTFDGVDVTGNRLAE 117
P H ++ V+G A W Y K L+ + V ++ + ++ + VD T ++ E
Sbjct: 239 PKHYVL-VHGACYGAWLW-YKLKPRLESAGHKVTVLDLAASGTNMKKIEDVD-TFSQYTE 409
Query: 118 EVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYER 154
++ ++ P +K+ + HSLGGL A+ K E+
Sbjct: 410 PLLQLMATIPPNKKVVLVGHSLGGLNIALAMEKFPEK 520
>TC213929 weakly similar to UP|Q94G63 (Q94G63) Ethylene-induced esterase,
partial (49%)
Length = 511
Score = 30.4 bits (67), Expect = 0.96
Identities = 24/96 (25%), Positives = 45/96 (46%), Gaps = 1/96 (1%)
Frame = +3
Query: 60 THLIIMVNGLIGSAHNWKYAAKQFLKRYPYDV-IVHCSECNSSTLTFDGVDVTGNRLAEE 118
T ++V+G A W Y K L+ + V +++ + + + VD T + E
Sbjct: 3 TRHYVLVHGACHGAWCW-YKLKPRLESEGHKVTVLNHAASGINMKKIEDVD-TFSEYTEP 176
Query: 119 VISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYER 154
++ ++ PS K+ + HSLGG+ A+ K E+
Sbjct: 177 LLQLLDTIPSNEKVVLVGHSLGGMSIAIAMEKFPEK 284
>TC218871
Length = 656
Score = 30.4 bits (67), Expect = 0.96
Identities = 24/82 (29%), Positives = 34/82 (41%)
Frame = -2
Query: 65 MVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVISVIK 124
++ +IG NW AK + KRYP I+ N+ FD + NR A +
Sbjct: 505 LIENVIGLLMNW*TTAKAYFKRYPKKRII----ANAPFFQFD----SSNRTA---LQA*T 359
Query: 125 RHPSVRKISFIAHSLGGLIARY 146
R S + LG +ARY
Sbjct: 358 RKASTTSHGHLTGGLGTSLARY 293
>TC218448 weakly similar to UP|DR29_ARATH (Q9SZA7) Probable disease resistance
protein At4g33300, partial (47%)
Length = 2012
Score = 30.0 bits (66), Expect = 1.2
Identities = 26/90 (28%), Positives = 40/90 (43%), Gaps = 17/90 (18%)
Frame = -1
Query: 92 IVHCSECNSSTLTFDGVDVTGNRLAEEVISVIKRHPSVRKIS-----------------F 134
I C +CN T F + G+ L +S+ +H ++ S F
Sbjct: 1754 ITSCRDCND-TADFGKYLIMGHSL----MSIFSKHTNLPISSGKHVRLTH*EISIYFNLF 1590
Query: 135 IAHSLGGLIARYAIAKLYERDISKELSQGN 164
++H LGG ++RY A Y+R IS +LS N
Sbjct: 1589 MSHMLGGSVSRYGQA--YKRRISSDLSLPN 1506
>AW235001
Length = 414
Score = 28.1 bits (61), Expect = 4.7
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +1
Query: 131 KISFIAHSLGGLIARYAIAK 150
K+SF+ HS+G LI R A+A+
Sbjct: 73 KLSFVGHSIGNLIIRAALAE 132
>TC222983
Length = 611
Score = 27.7 bits (60), Expect = 6.2
Identities = 9/20 (45%), Positives = 15/20 (75%)
Frame = +3
Query: 57 SNPTHLIIMVNGLIGSAHNW 76
S+ HL++MVNG++G +W
Sbjct: 552 SSADHLVVMVNGILGRETDW 611
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,467,445
Number of Sequences: 63676
Number of extensions: 169582
Number of successful extensions: 804
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 798
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 801
length of query: 285
length of database: 12,639,632
effective HSP length: 96
effective length of query: 189
effective length of database: 6,526,736
effective search space: 1233553104
effective search space used: 1233553104
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146940.1


