

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146914.10 + phase: 0
(541 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
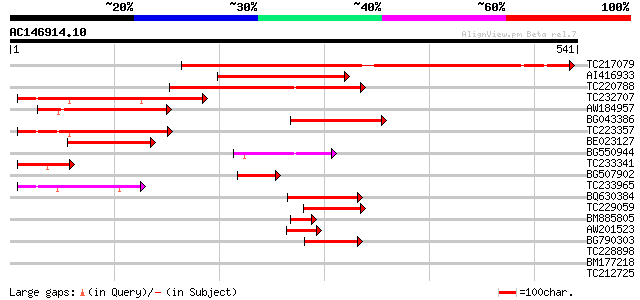
Score E
Sequences producing significant alignments: (bits) Value
TC217079 similar to UP|Q9FJI9 (Q9FJI9) Similarity to calmodulin-... 568 e-162
AI416933 similar to PIR|T03793|T03 calmodulin-binding protein TC... 155 3e-38
TC220788 weakly similar to UP|Q9FKL6 (Q9FKL6) Calmodulin-binding... 145 6e-35
TC232707 143 2e-34
AW184957 similar to GP|9758319|dbj calmodulin-binding protein {A... 127 1e-29
BG043386 127 1e-29
TC223357 similar to UP|Q9FKL6 (Q9FKL6) Calmodulin-binding protei... 127 2e-29
BE023127 80 2e-15
BG550944 77 1e-14
TC233341 similar to UP|Q8PWH4 (Q8PWH4) Conserved protein, partia... 74 1e-13
BG507902 72 6e-13
TC233965 66 3e-11
BQ630384 62 8e-10
TC229059 similar to UP|Q9M451 (Q9M451) Calmodulin-binding protei... 54 2e-07
BM885805 54 2e-07
AW201523 similar to PIR|T03793|T037 calmodulin-binding protein T... 47 3e-05
BG790303 similar to GP|24943204|gb calmodulin-binding protein 60... 42 5e-04
TC228898 30 2.1
BM177218 30 2.1
TC212725 29 6.1
>TC217079 similar to UP|Q9FJI9 (Q9FJI9) Similarity to calmodulin-binding
protein, partial (26%)
Length = 1412
Score = 568 bits (1464), Expect = e-162
Identities = 293/376 (77%), Positives = 319/376 (83%), Gaps = 1/376 (0%)
Frame = +2
Query: 165 MVGEISYTDNSSWTRSRRFRLGVRVVDNFDGIRIREAKTDSFIVRDHRGELYKKHHPPSL 224
MVGEISYTDNSSWTRSRRFRLG RVVDNFDG+ IREAKT+SFIVRDHRGELYKKHHPPSL
Sbjct: 2 MVGEISYTDNSSWTRSRRFRLGARVVDNFDGVGIREAKTESFIVRDHRGELYKKHHPPSL 181
Query: 225 SDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVE 284
SDEVWRLEKIGKDGAFH+RLSREKI TV++FLTLLNLDPAKLR+ILGTGMSAKMWEVTVE
Sbjct: 182 SDEVWRLEKIGKDGAFHKRLSREKILTVREFLTLLNLDPAKLRSILGTGMSAKMWEVTVE 361
Query: 285 HARTCVLDTTRHVSFASHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFISDHPL 344
HARTCVLDTTRHV F S+SQ+P VVFNAVG+VTGLL+E EYV VDKL+ETEK
Sbjct: 362 HARTCVLDTTRHVYFPSNSQEPGVVFNAVGQVTGLLSECEYVTVDKLTETEK-------- 517
Query: 345 VSLADAQISVISALNQCD-FASFEDEVSLMDGYSHLTNVHYSPSSPRTEGSSANKLLALQ 403
ADAQ SV +AL Q + + +FEDE SLMDG SHLTNV YSPSSP+TEGSSANK+LA Q
Sbjct: 518 ---ADAQNSVTAALRQGEKYTTFEDEDSLMDGSSHLTNVLYSPSSPKTEGSSANKILAPQ 688
Query: 404 KTGGFNYTQESASSTDIMPSIYSVGGTSSLDDYGLPNFESLGLRYDQHLGFPVQVSNSLI 463
KTGGFNY SASS DIM SIYSVGGTSSLDDY LPNF+S+G RYDQ L FPVQVSNSLI
Sbjct: 689 KTGGFNYPPASASSPDIMSSIYSVGGTSSLDDYCLPNFDSMGFRYDQTLSFPVQVSNSLI 868
Query: 464 CDMDSIVHAFGDEDHLQFFDADLQSQCHIEADLHSAVDSFMPVSSTSMTKGKAQRRWRKV 523
CD DS+ HAF DEDHLQFFD DLQS H++ADL A+DSF T+ G AQRRWRKV
Sbjct: 869 CDTDSMAHAFSDEDHLQFFDTDLQS--HVQADLQGAIDSFRLARPTA--NGGAQRRWRKV 1036
Query: 524 VNVLKWFMVKKRRNQL 539
NVLKWFMV+KR NQ+
Sbjct: 1037CNVLKWFMVRKRGNQI 1084
>AI416933 similar to PIR|T03793|T03 calmodulin-binding protein TCB60 - common
tobacco, partial (22%)
Length = 381
Score = 155 bits (393), Expect = 3e-38
Identities = 72/126 (57%), Positives = 98/126 (77%)
Frame = +2
Query: 199 REAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLTL 258
REAKT++F V+DHRGELYKKH+PP+L+DEVWRLEKIGKDG+FH+RL++ I TV+D + L
Sbjct: 2 REAKTEAFTVKDHRGELYKKHYPPALNDEVWRLEKIGKDGSFHKRLNKAGIYTVEDVVQL 181
Query: 259 LNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEVTG 318
+ DP +LR ILG+GMS KMW+V VEHA+TCVL +V + ++ VVFN + E++G
Sbjct: 182 VVRDPQRLRNILGSGMSNKMWDVLVEHAKTCVLSGKLYVYYPDDARNVGVVFNNIYELSG 361
Query: 319 LLAESE 324
L+ +
Sbjct: 362 LITNDQ 379
>TC220788 weakly similar to UP|Q9FKL6 (Q9FKL6) Calmodulin-binding protein,
partial (13%)
Length = 1435
Score = 145 bits (365), Expect = 6e-35
Identities = 74/189 (39%), Positives = 117/189 (61%), Gaps = 2/189 (1%)
Frame = +2
Query: 153 GDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDNFD-GIRIREAKTDSFIVRDH 211
GD + LK+G+ V ++ +TDNS W RSR+FRLG +VV I+E +++ F+V+D+
Sbjct: 5 GDRFITLKNGVGCVNKLVFTDNSRWIRSRKFRLGAKVVPPISIEANIKEGRSEPFVVKDY 184
Query: 212 RGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILG 271
RGE + KHHPPSL+D+VWRLE+I KDG H RLS I TV+D L L +P+ L +G
Sbjct: 185 RGEAF*KHHPPSLNDDVWRLEQIAKDGKIHDRLSLHGIHTVQDLLRLYTTNPSSLLEKVG 364
Query: 272 TGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEVTGLLAESE-YVAVDK 330
++ + W +EHA+TC +D + + Q ++FN++ + G+ + + Y++ D
Sbjct: 365 -NITKRSWITIIEHAKTCAIDDDETFVYHTAEQSIGLLFNSIYILVGVTFDGQNYLSPDI 541
Query: 331 LSETEKAFI 339
L+ EK +
Sbjct: 542 LNPNEKHLV 568
>TC232707
Length = 764
Score = 143 bits (361), Expect = 2e-34
Identities = 76/186 (40%), Positives = 123/186 (65%), Gaps = 5/186 (2%)
Frame = +2
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCG---KEMNTSESR 64
SV+ EV+ ++++ NL +EPL++RVV EEV+ A+++ S ++ + M+ +
Sbjct: 209 SVIGEVVMVKNLENLFSA-MEPLLKRVVGEEVDQAMRQWSRSFARSPSLRLQAMDQQQPS 385
Query: 65 TLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDF 124
TLQL F +SLP+FTG+RI DG+ + I L+D G+ V T ++ K+EIVV++GDF
Sbjct: 386 TLQLCFSKRLSLPIFTGSRILDVDGNPINIVLMDKSNGQGVPTSLSNAIKLEIVVVDGDF 565
Query: 125 --EEESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRR 182
+ + W E+F +IV+ER+GK+PLL G++ + ++DG+ G+I +TDNSSW R R+
Sbjct: 566 PLNDNDEDWTSEEFNRHIVKERNGKRPLLAGELNVIMRDGIAPTGDIEFTDNSSWIRCRK 745
Query: 183 FRLGVR 188
FR+ VR
Sbjct: 746 FRVAVR 763
>AW184957 similar to GP|9758319|dbj calmodulin-binding protein {Arabidopsis
thaliana}, partial (21%)
Length = 424
Score = 127 bits (320), Expect = 1e-29
Identities = 66/133 (49%), Positives = 96/133 (71%), Gaps = 5/133 (3%)
Frame = +3
Query: 27 LEPLVRRVVREEVELALKK----HLSSIKQTCGKEMNTSESRTLQLQFENSISLPVFTGA 82
LEP++RRVV EEVE AL K LS ++ K + + R+LQL+F + +SLP+FTG
Sbjct: 30 LEPILRRVVSEEVERALAKLGPARLSG--RSPPKMIEGPDGRSLQLKFRSRLSLPLFTGG 203
Query: 83 RIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESD-IWMPEDFKNNIV 141
++EGE G+ + + L+DA +G +V +GPES K+++VVLEGDF E D W EDF++++V
Sbjct: 204 KVEGEQGAPIHVVLIDANSGSIVTSGPESCVKLDVVVLEGDFNNEDDEDWTQEDFESHVV 383
Query: 142 RERDGKKPLLTGD 154
+ER+GK+PLLTGD
Sbjct: 384 KEREGKRPLLTGD 422
>BG043386
Length = 417
Score = 127 bits (319), Expect = 1e-29
Identities = 65/91 (71%), Positives = 73/91 (79%)
Frame = +2
Query: 269 ILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEVTGLLAESEYVAV 328
ILGTGMSAKMWEVTVEHARTCVLDTTRHV F S+SQ+P VVFNAVG+VTGLL+E +YV V
Sbjct: 143 ILGTGMSAKMWEVTVEHARTCVLDTTRHVYFPSNSQEPGVVFNAVGQVTGLLSECDYVTV 322
Query: 329 DKLSETEKAFISDHPLVSLADAQISVISALN 359
DKL+ETEKAFI L+ L S S +N
Sbjct: 323 DKLTETEKAFIPHPLLLCLLFPSFSYFSLIN 415
>TC223357 similar to UP|Q9FKL6 (Q9FKL6) Calmodulin-binding protein, partial
(25%)
Length = 689
Score = 127 bits (318), Expect = 2e-29
Identities = 69/153 (45%), Positives = 103/153 (67%), Gaps = 5/153 (3%)
Frame = +2
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCG----KEMNTSES 63
SV+ E +K+ S++ L LEP++RRVV EEVE AL K L K G K + +
Sbjct: 227 SVIVEALKVDSLQKLCSS-LEPILRRVVSEEVERALAK-LGPAKLNTGRSSPKRIEGPDG 400
Query: 64 RTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGD 123
+ LQL F+ +SLP+FTG ++EGE G+ + I L+DA + +V +GPES +++++VLEGD
Sbjct: 401 KNLQLHFKTRLSLPLFTGGKVEGEQGTAIHIVLIDANSDHIVTSGPESCVRLDVIVLEGD 580
Query: 124 F-EEESDIWMPEDFKNNIVRERDGKKPLLTGDV 155
F E+ D W E+ ++IV+ER+GK+PLLTGD+
Sbjct: 581 FNNEDDDNWDEEEVDSHIVKEREGKRPLLTGDL 679
>BE023127
Length = 277
Score = 80.1 bits (196), Expect = 2e-15
Identities = 39/85 (45%), Positives = 55/85 (63%), Gaps = 1/85 (1%)
Frame = +3
Query: 56 KEMNTSESRTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKV 115
K + + +LQLQF +SLP+FTG ++EGE GS + I L+D TG VV GP S K+
Sbjct: 21 KGIEGPDDSSLQLQFRTRLSLPLFTGGKVEGEHGSAIHIVLIDTTTGHVVTCGPASCVKL 200
Query: 116 EIVVLEGDF-EEESDIWMPEDFKNN 139
+++VLEGDF E+ D W E F ++
Sbjct: 201 DVIVLEGDFNNEDDDNWSEEYFDSH 275
>BG550944
Length = 414
Score = 77.4 bits (189), Expect = 1e-14
Identities = 40/101 (39%), Positives = 59/101 (57%), Gaps = 2/101 (1%)
Frame = +1
Query: 214 ELYKKHHPP--SLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILG 271
+ YKKH+PP L+D++WRL+KI K+G H++LS I VKD L L + L + G
Sbjct: 115 QAYKKHYPPYLKLNDDIWRLKKIAKEGKIHKQLSSRGIHNVKDLLRLYITNEPSLYEMFG 294
Query: 272 TGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNA 312
+ K W V EHA+ CV+D + S+ S Q ++FN+
Sbjct: 295 -NIPKKSWLVITEHAKACVIDDYQLYSYHSQELQIGLLFNS 414
>TC233341 similar to UP|Q8PWH4 (Q8PWH4) Conserved protein, partial (7%)
Length = 714
Score = 74.3 bits (181), Expect = 1e-13
Identities = 41/75 (54%), Positives = 52/75 (68%), Gaps = 20/75 (26%)
Frame = +2
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRV--------------------VREEVELALKKHL 47
SVVR+VMKLQS+ +L+EPILEPLVR+V V+EEVE ALK+HL
Sbjct: 485 SVVRDVMKLQSLGHLLEPILEPLVRKVAADNLCCIQFMVPIVFA*LQVKEEVEAALKRHL 664
Query: 48 SSIKQTCGKEMNTSE 62
+S+KQTCGKE +T+E
Sbjct: 665 TSMKQTCGKEFHTTE 709
>BG507902
Length = 134
Score = 72.0 bits (175), Expect = 6e-13
Identities = 31/41 (75%), Positives = 37/41 (89%)
Frame = +1
Query: 218 KHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLTL 258
KHHPP L+DEVWRLEKIGKDGAFH++LS+E I +V+DFL L
Sbjct: 1 KHHPPMLNDEVWRLEKIGKDGAFHKKLSKEGINSVQDFLKL 123
>TC233965
Length = 565
Score = 66.2 bits (160), Expect = 3e-11
Identities = 44/131 (33%), Positives = 71/131 (53%), Gaps = 9/131 (6%)
Frame = +2
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALK-----KHLSSIKQTCGKEMNTSE 62
SV+ EV+ +++++NL LEPL+RRVV EEVE ++ + +S + + +
Sbjct: 173 SVIGEVVMVKNLQNLFSG-LEPLLRRVVNEEVERVMRHCTVPRTISRSPSLRIEAASLEK 349
Query: 63 SRTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGK----VVCTGPESSAKVEIV 118
+L F + +P+FTG+RI DG+ +++ LVD G V T K+EIV
Sbjct: 350 PSNYELMFSKKLLVPIFTGSRIVDIDGNPIQVILVDKSGGDGELVAVPTSVPQPIKLEIV 529
Query: 119 VLEGDFEEESD 129
VL+GDF +
Sbjct: 530 VLDGDFPNNKE 562
>BQ630384
Length = 429
Score = 61.6 bits (148), Expect = 8e-10
Identities = 27/71 (38%), Positives = 46/71 (64%)
Frame = +2
Query: 266 LRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEVTGLLAESEY 325
L ILG+GMS +MWE TVEHA+TCVL V + + ++FN + E+ GL+++ ++
Sbjct: 14 LTQILGSGMSNRMWENTVEHAKTCVLGGKLFVYYIDETNSSGIMFNNIYELRGLISDGQF 193
Query: 326 VAVDKLSETEK 336
+++ L+ +K
Sbjct: 194 FSLESLTPNQK 226
>TC229059 similar to UP|Q9M451 (Q9M451) Calmodulin-binding protein
(Fragment), partial (89%)
Length = 1424
Score = 53.5 bits (127), Expect = 2e-07
Identities = 23/59 (38%), Positives = 38/59 (63%)
Frame = +1
Query: 281 VTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFI 339
+ VEHA+TCVL +V + ++ VVFN + E++GL+A +Y + D LSE +K ++
Sbjct: 7 ILVEHAKTCVLSGKLYVYYPEDARNVGVVFNNIYELSGLIANDQYYSADSLSENQKVYV 183
>BM885805
Length = 421
Score = 53.5 bits (127), Expect = 2e-07
Identities = 24/24 (100%), Positives = 24/24 (100%)
Frame = +2
Query: 269 ILGTGMSAKMWEVTVEHARTCVLD 292
ILGTGMSAKMWEVTVEHARTCVLD
Sbjct: 350 ILGTGMSAKMWEVTVEHARTCVLD 421
>AW201523 similar to PIR|T03793|T037 calmodulin-binding protein TCB60 -
common tobacco, partial (4%)
Length = 406
Score = 46.6 bits (109), Expect = 3e-05
Identities = 20/33 (60%), Positives = 24/33 (72%)
Frame = +1
Query: 265 KLRTILGTGMSAKMWEVTVEHARTCVLDTTRHV 297
KLR ILG GMS KMWEVT++HA TC + H+
Sbjct: 13 KLRNILGIGMSDKMWEVTIKHAMTCDTGSKMHI 111
>BG790303 similar to GP|24943204|gb calmodulin-binding protein 60-D
{Phaseolus vulgaris}, partial (41%)
Length = 398
Score = 42.4 bits (98), Expect = 5e-04
Identities = 17/55 (30%), Positives = 34/55 (60%)
Frame = +3
Query: 282 TVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEK 336
TVEHA+TCVL V + + ++FN + E+ GL+++ ++ +++ L+ +K
Sbjct: 3 TVEHAKTCVLGGKLFVYYIDETNSSGIMFNNIYELRGLISDGQFFSLESLTPNQK 167
>TC228898
Length = 920
Score = 30.4 bits (67), Expect = 2.1
Identities = 37/172 (21%), Positives = 68/172 (39%), Gaps = 24/172 (13%)
Frame = +3
Query: 277 KMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAV-----GEVTG------------- 318
KMW+VT++HA+TC ++ ++ H+ V N++ ++ G
Sbjct: 3 KMWDVTIKHAKTCE-KGNKYYAYRGHNFT--VFLNSICQLVRADINGQTFPGRELSNMTR 173
Query: 319 ----LLAESEYVAVDKLSETEKAFISD-HPLVSLADAQISVISALNQCDFASFEDEVSLM 373
L Y + L E + A ++ L + + + A +Q D+ F D+ + +
Sbjct: 174 SYMEKLVREAYARWNDLEEIDAALLTQGETLEEFPNNHQASLIAYDQNDY--FGDKSAEV 347
Query: 374 DGYSHLTNVHYSPSSPRTEGSSANKLLALQKTGGFNYT-QESASSTDIMPSI 424
Y N S + G+ GG Y+ +S S +DI PS+
Sbjct: 348 GNYVPTHNAQMGCSDWQVNGTFGTTSFV---NGGIPYSFSDSQSDSDITPSV 494
>BM177218
Length = 437
Score = 30.4 bits (67), Expect = 2.1
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 8/63 (12%)
Frame = +3
Query: 16 LQSVRNLMEPI--------LEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTLQ 67
L +RN+M+ + E +RRVVREEVE ++ LSS + +++TS + +
Sbjct: 231 LSGLRNVMKGLCTNDCELHFERFLRRVVREEVECKIQDFLSS--RGWVNQISTSRATPFE 404
Query: 68 LQF 70
L+F
Sbjct: 405 LRF 413
>TC212725
Length = 415
Score = 28.9 bits (63), Expect = 6.1
Identities = 12/28 (42%), Positives = 17/28 (59%)
Frame = +3
Query: 382 VHYSPSSPRTEGSSANKLLALQKTGGFN 409
VH+SPS P T G+ K +A K G ++
Sbjct: 258 VHWSPSVPSTSGTLHRKTIASAKNGSYD 341
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.134 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,619,140
Number of Sequences: 63676
Number of extensions: 287915
Number of successful extensions: 1456
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1430
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1439
length of query: 541
length of database: 12,639,632
effective HSP length: 102
effective length of query: 439
effective length of database: 6,144,680
effective search space: 2697514520
effective search space used: 2697514520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146914.10


