

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.4 + phase: 0
(321 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
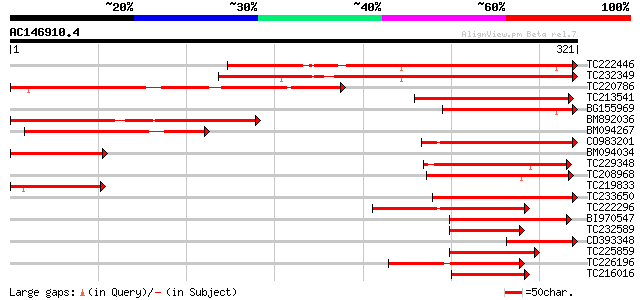
Score E
Sequences producing significant alignments: (bits) Value
TC222446 similar to UP|O81241 (O81241) Dc3 promoter-binding fact... 233 9e-62
TC232349 similar to UP|O81241 (O81241) Dc3 promoter-binding fact... 183 1e-46
TC220786 similar to UP|Q9C5Q3 (Q9C5Q3) BZIP protein DPBF3, parti... 179 1e-45
TC213541 similar to UP|Q9LES3 (Q9LES3) Promoter-binding factor-l... 148 4e-36
BG155969 similar to PIR|T12622|T126 Dc3 promoter-binding factor-... 127 7e-30
BM892036 119 2e-27
BM094267 weakly similar to GP|13346155|gb| bZIP protein DPBF3 {A... 97 1e-20
CO983201 93 2e-19
BM094034 weakly similar to GP|11994451|dbj abscisic acid respons... 93 2e-19
TC229348 homologue to UP|Q94KA6 (Q94KA6) BZIP transcription fact... 82 2e-16
TC208968 similar to UP|GBF4_ARATH (P42777) G-box binding factor ... 75 3e-14
TC219833 similar to UP|Q9C5Q3 (Q9C5Q3) BZIP protein DPBF3, parti... 69 3e-12
TC233650 similar to UP|Q84JC9 (Q84JC9) BZIP transcription factor... 66 2e-11
TC222296 similar to UP|P93426 (P93426) Leucine zipper protein, p... 64 1e-10
BI970547 weakly similar to GP|29027731|dbj bZIP transcription fa... 62 3e-10
TC232589 similar to UP|Q8LK78 (Q8LK78) ABA response element bind... 57 1e-08
CD393348 weakly similar to PIR|T12622|T126 Dc3 promoter-binding ... 51 6e-07
TC225859 similar to UP|O22677 (O22677) BZIP DNA-binding protein,... 50 2e-06
TC226196 UP|Q8L5W2 (Q8L5W2) BZIP transcription factor ATB2, comp... 49 2e-06
TC216016 similar to UP|P93839 (P93839) G/HBF-1, partial (61%) 48 5e-06
>TC222446 similar to UP|O81241 (O81241) Dc3 promoter-binding factor-3
(Fragment), partial (46%)
Length = 765
Score = 233 bits (594), Expect = 9e-62
Identities = 137/203 (67%), Positives = 152/203 (74%), Gaps = 5/203 (2%)
Frame = +3
Query: 124 EKQQTLGEMTLEDFLVKAGVVGESFHG-KESGLLRVDSNEDSRQKVSHGLHWMQYPVHSV 182
E+Q TLGEM LEDFLVKAGV+ E+ K+ + VDSN S Q HG HW+QY
Sbjct: 3 ERQATLGEMXLEDFLVKAGVIAEALPTTKDRAMSGVDSNGASSQ---HG-HWLQYQQLPS 170
Query: 183 QQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAAIS--PSSLMGTLSDTQTLGRKRVA 240
QQ + M G+ A HAIQQPFQV N LDAA S P+SL G LSDTQTLGRKR
Sbjct: 171 SVQQ----PNVMGGYVAGHAIQQPFQVGVNLVLDAAYSETPASLKGALSDTQTLGRKRGV 338
Query: 241 SGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEKE 300
SGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELE+KVSRLEEENERLRR NEME+
Sbjct: 339 SGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELEIKVSRLEEENERLRRLNEMERA 518
Query: 301 VPTAPPPE--PKNQLRRTNSASF 321
+P+ PPPE PK+QLRRT+SA F
Sbjct: 519 LPSVPPPEPKPKHQLRRTSSAIF 587
>TC232349 similar to UP|O81241 (O81241) Dc3 promoter-binding factor-3
(Fragment), partial (50%)
Length = 834
Score = 183 bits (464), Expect = 1e-46
Identities = 109/212 (51%), Positives = 138/212 (64%), Gaps = 9/212 (4%)
Frame = +3
Query: 119 DRKSREKQQTLGEMTLEDFLVKAGVVGESFHGKE--SGLLRVDSNEDSRQKVSHGLHWMQ 176
++K +++ LGEMTLEDF AGVV + + S + VDSN Q S W+Q
Sbjct: 6 EKKFQDRXPXLGEMTLEDFXXXAGVVAGAXXNRTNXSTIAGVDSNVAVPQFXSQA-QWIQ 182
Query: 177 YPVHSVQQQQHQYEKHTMPG-FAAVHAIQQPFQVAGNQALDAAIS------PSSLMGTLS 229
YP Q Q+Q+ ++ G + + QP + +LD + + PSSLMGT+S
Sbjct: 183 YP-----QAQYQHPPQSLMGMYMPSQGMVQPLHMGAGASLDVSFADNQMAMPSSLMGTMS 347
Query: 230 DTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENE 289
DTQT GRK+ S ++EKTVERRQKRMIKNRESAARSRAR+QAYT ELE KVSRLEEENE
Sbjct: 348 DTQTPGRKKSTSEDMIEKTVERRQKRMIKNRESAARSRARKQAYTNELENKVSRLEEENE 527
Query: 290 RLRRQNEMEKEVPTAPPPEPKNQLRRTNSASF 321
RLR++ E+E+ + APPPEPK QLRR SA F
Sbjct: 528 RLRKRKELEQMLSCAPPPEPKYQLRRIASAPF 623
>TC220786 similar to UP|Q9C5Q3 (Q9C5Q3) BZIP protein DPBF3, partial (29%)
Length = 1165
Score = 179 bits (454), Expect = 1e-45
Identities = 104/192 (54%), Positives = 123/192 (63%), Gaps = 2/192 (1%)
Frame = +3
Query: 1 MGSQGGTVS--ESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEA 58
MGSQGGT E KT L+R GSLYNLTLDEVQN LGNLG+P SMNLDELLKSVW+ E+
Sbjct: 633 MGSQGGTTQDQEPKTGSLTRQGSLYNLTLDEVQNQLGNLGEPFRSMNLDELLKSVWTAES 812
Query: 59 GEVSDFGGSDVAATAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKKRGVDR 118
G + A+AG ++ N Q SLTLSG+LSKKT+DEVW+DMQ K
Sbjct: 813 GTDAYMQHGGQVASAGSSL--------NPQGSLTLSGNLSKKTIDEVWRDMQQNK----- 953
Query: 119 DRKSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLRVDSNEDSRQKVSHGLHWMQYP 178
+E+Q TLGEMTLEDFLVKAGV E F ++ + S DS+ S HWMQY
Sbjct: 954 -SVGKERQPTLGEMTLEDFLVKAGVATEPFPNEDGAMAM--SGVDSQHNTSQHAHWMQYQ 1124
Query: 179 VHSVQQQQHQYE 190
+ SVQQQ Q +
Sbjct: 1125LTSVQQQPQQQQ 1160
>TC213541 similar to UP|Q9LES3 (Q9LES3) Promoter-binding factor-like protein
(ABA-responsive element binding protein 3) (AREB3),
partial (31%)
Length = 741
Score = 148 bits (373), Expect = 4e-36
Identities = 74/90 (82%), Positives = 85/90 (94%)
Frame = +1
Query: 230 DTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENE 289
D+QT GRKR ASG VVEK VERRQKRMIKNRESAARSRAR+QAYTQELE+KVS+LEEENE
Sbjct: 1 DSQTAGRKRDASGNVVEKIVERRQKRMIKNRESAARSRARKQAYTQELEIKVSQLEEENE 180
Query: 290 RLRRQNEMEKEVPTAPPPEPKNQLRRTNSA 319
RLRRQNE+E+ +P+APPP+PK+QLRRT+SA
Sbjct: 181 RLRRQNEIERALPSAPPPDPKHQLRRTSSA 270
>BG155969 similar to PIR|T12622|T126 Dc3 promoter-binding factor-2 - common
sunflower, partial (43%)
Length = 414
Score = 127 bits (319), Expect = 7e-30
Identities = 66/78 (84%), Positives = 71/78 (90%), Gaps = 2/78 (2%)
Frame = +2
Query: 246 EKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEKEVPTAP 305
EKTVERRQKRMIKNRESAARSRARRQAYTQELE+KVSRLEEENERLRR NEME+ +P+ P
Sbjct: 2 EKTVERRQKRMIKNRESAARSRARRQAYTQELEIKVSRLEEENERLRRLNEMERALPSVP 181
Query: 306 PPE--PKNQLRRTNSASF 321
PPE PK QLRRT+SA F
Sbjct: 182 PPEPKPKQQLRRTSSAIF 235
>BM892036
Length = 422
Score = 119 bits (298), Expect = 2e-27
Identities = 72/144 (50%), Positives = 93/144 (64%), Gaps = 2/144 (1%)
Frame = +3
Query: 1 MGSQG-GTVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAG 59
+GS G G S + LSR GS Y+LTLDEV LG++GKPLGSMNLDELL++VW+ EA
Sbjct: 9 VGSLGNGQQSHLQPSSLSRQGSWYSLTLDEVNCQLGDMGKPLGSMNLDELLQNVWTAEAS 188
Query: 60 EVSDFGGSDVAATAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDM-QGKKRGVDR 118
+ S V NM + Q SLTL+ LS KTVD+VW+++ QG+K+
Sbjct: 189 K-----SSVVVGVESENMSSSS-SSLQRQASLTLARALSGKTVDDVWREIQQGQKKKYGE 350
Query: 119 DRKSREKQQTLGEMTLEDFLVKAG 142
D +S+E + TLGE TLEDFLV+AG
Sbjct: 351 DVRSQEGEMTLGETTLEDFLVQAG 422
>BM094267 weakly similar to GP|13346155|gb| bZIP protein DPBF3 {Arabidopsis
thaliana}, partial (25%)
Length = 428
Score = 97.1 bits (240), Expect = 1e-20
Identities = 52/105 (49%), Positives = 67/105 (63%)
Frame = +3
Query: 9 SESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWSVEAGEVSDFGGSD 68
SE+ + R S+Y+LTLDEVQN LG+LGKPL SMNLDELLK+VW+ EA +
Sbjct: 126 SENW*MGTQRQNSMYSLTLDEVQNQLGDLGKPLTSMNLDELLKNVWTAEASQTIGMDNEG 305
Query: 69 VAATAGGNMQHNQLGGFNSQESLTLSGDLSKKTVDEVWKDMQGKK 113
+ + +QH Q SL+L+G LSK TVDEVW+D+Q K
Sbjct: 306 TSQASQAALQH--------QASLSLTGALSKMTVDEVWRDIQENK 416
>CO983201
Length = 774
Score = 92.8 bits (229), Expect = 2e-19
Identities = 53/88 (60%), Positives = 62/88 (70%)
Frame = -3
Query: 234 LGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRR 293
LGRKR EKT+ERR +R IKNRE AARSRAR+QAY EL KVSRLEEEN +L++
Sbjct: 763 LGRKRDXXD-AYEKTLERRLRRKIKNREXAARSRARKQAYHNELVGKVSRLEEENVKLKK 587
Query: 294 QNEMEKEVPTAPPPEPKNQLRRTNSASF 321
+ E E+ + P PE K QLRR NSA F
Sbjct: 586 EKEFEERLLPDPLPERKYQLRRHNSAFF 503
>BM094034 weakly similar to GP|11994451|dbj abscisic acid responsive
elements-binding factor {Arabidopsis thaliana}, partial
(9%)
Length = 433
Score = 92.8 bits (229), Expect = 2e-19
Identities = 47/56 (83%), Positives = 48/56 (84%), Gaps = 1/56 (1%)
Frame = +1
Query: 1 MGSQGGTVSESKTL-PLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVWS 55
MGSQGG V E KT PL+R GSLYNLTLDEV N LGNLGKPLGSMNLDELLKSVWS
Sbjct: 265 MGSQGGAVQEPKTTTPLARQGSLYNLTLDEVHNQLGNLGKPLGSMNLDELLKSVWS 432
>TC229348 homologue to UP|Q94KA6 (Q94KA6) BZIP transcription factor 6,
partial (34%)
Length = 786
Score = 82.4 bits (202), Expect = 2e-16
Identities = 48/92 (52%), Positives = 61/92 (66%), Gaps = 8/92 (8%)
Frame = +1
Query: 235 GRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRR- 293
GRK SG VEK +ERRQ+RMIKNRESAARSRAR+QAYT ELE +V++L+EENE L++
Sbjct: 205 GRK---SGGAVEKVIERRQRRMIKNRESAARSRARKQAYTMELEAEVAKLKEENEELQKK 375
Query: 294 -------QNEMEKEVPTAPPPEPKNQLRRTNS 318
Q KE+ + +LRRT +
Sbjct: 376 QAEIMEIQKNQVKEMMNLQREVKRRRLRRTQT 471
>TC208968 similar to UP|GBF4_ARATH (P42777) G-box binding factor 4
(AtbZIP40), partial (27%)
Length = 1199
Score = 75.5 bits (184), Expect = 3e-14
Identities = 45/93 (48%), Positives = 57/93 (60%), Gaps = 10/93 (10%)
Frame = +2
Query: 237 KRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEEN-------- 288
KR A V+K ++Q+RMIKNRESAARSR R+QAYT ELE V +LE+EN
Sbjct: 497 KRRAVEEPVDKATLQKQRRMIKNRESAARSRERKQAYTSELEYLVHQLEQENVQLLNEEA 676
Query: 289 --ERLRRQNEMEKEVPTAPPPEPKNQLRRTNSA 319
R R++ E +P P+PK +LRR NSA
Sbjct: 677 EMRRQRKKQLFECIIPIEVMPKPKKKLRRVNSA 775
>TC219833 similar to UP|Q9C5Q3 (Q9C5Q3) BZIP protein DPBF3, partial (13%)
Length = 526
Score = 68.9 bits (167), Expect = 3e-12
Identities = 35/58 (60%), Positives = 42/58 (72%), Gaps = 4/58 (6%)
Frame = +3
Query: 1 MGSQGG----TVSESKTLPLSRSGSLYNLTLDEVQNHLGNLGKPLGSMNLDELLKSVW 54
MG+ GG +S PL R S+Y+LTLDEVQN LG+LGKPL SMN+DELLK+VW
Sbjct: 351 MGTHGGGGDSNGKQSPLQPLVRQNSMYSLTLDEVQNQLGDLGKPLTSMNIDELLKNVW 524
>TC233650 similar to UP|Q84JC9 (Q84JC9) BZIP transcription factor, partial
(17%)
Length = 792
Score = 65.9 bits (159), Expect = 2e-11
Identities = 40/85 (47%), Positives = 59/85 (69%), Gaps = 3/85 (3%)
Frame = +3
Query: 240 ASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQ-NEME 298
ASG +++R R+IKNRESA RSRAR+QAY + LE++++RL EEN RL+RQ E++
Sbjct: 303 ASGSKKTTLLDQRHARIIKNRESAVRSRARKQAYRKGLEVEIARLTEENSRLKRQLKELQ 482
Query: 299 KEVPTAP-PPEPK-NQLRRTNSASF 321
+ ++ PP P+ L RT+S+ F
Sbjct: 483 CCLSSSDNPPTPRMAALCRTSSSPF 557
>TC222296 similar to UP|P93426 (P93426) Leucine zipper protein, partial (24%)
Length = 945
Score = 63.5 bits (153), Expect = 1e-10
Identities = 36/89 (40%), Positives = 54/89 (60%)
Frame = +3
Query: 206 PFQVAGNQALDAAISPSSLMGTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAAR 265
PF G+ + + + + S++ G++R V+K ++ +RMIKNRESAAR
Sbjct: 360 PFPAEGSSSSVEPFANNGVGSAPSNSVQKGKRRAVEE-PVDKATLQKLRRMIKNRESAAR 536
Query: 266 SRARRQAYTQELELKVSRLEEENERLRRQ 294
SR R+QAYT ELE V +LE+EN RL ++
Sbjct: 537 SRERKQAYTSELEYLVHQLEQENARLLKE 623
>BI970547 weakly similar to GP|29027731|dbj bZIP transcription factor
{Arabidopsis thaliana}, partial (15%)
Length = 697
Score = 62.4 bits (150), Expect = 3e-10
Identities = 33/70 (47%), Positives = 50/70 (71%), Gaps = 1/70 (1%)
Frame = -1
Query: 250 ERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQ-NEMEKEVPTAPPPE 308
++R R+IKNRESA RSRAR+QAY + LE+++SRL EEN RL+RQ E+++ + ++ P
Sbjct: 493 DQRHTRVIKNRESAVRSRARKQAYRKGLEVEISRLTEENSRLKRQLKELQRCLCSSHTPR 314
Query: 309 PKNQLRRTNS 318
R ++S
Sbjct: 313 MAAPCRTSSS 284
>TC232589 similar to UP|Q8LK78 (Q8LK78) ABA response element binding factor,
partial (10%)
Length = 464
Score = 57.0 bits (136), Expect = 1e-08
Identities = 27/42 (64%), Positives = 35/42 (83%)
Frame = +2
Query: 250 ERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERL 291
++R R+IKNRESA RSRAR+QAY + LE+++SRL EEN RL
Sbjct: 338 DQRHTRVIKNRESAVRSRARKQAYRKGLEVEISRLTEENSRL 463
>CD393348 weakly similar to PIR|T12622|T126 Dc3 promoter-binding factor-2 -
common sunflower, partial (22%)
Length = 365
Score = 51.2 bits (121), Expect = 6e-07
Identities = 25/40 (62%), Positives = 29/40 (72%)
Frame = -1
Query: 282 SRLEEENERLRRQNEMEKEVPTAPPPEPKNQLRRTNSASF 321
+R EEENERLR+ E+E +P PPPEPK QLRR SA F
Sbjct: 365 ARGEEENERLRKLKELELMLPCEPPPEPKYQLRRIASAPF 246
>TC225859 similar to UP|O22677 (O22677) BZIP DNA-binding protein, partial
(59%)
Length = 1132
Score = 49.7 bits (117), Expect = 2e-06
Identities = 24/51 (47%), Positives = 36/51 (70%)
Frame = +1
Query: 250 ERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQNEMEKE 300
ER++KRM+ NRESA RSR R+Q ++L +VSRL+ N++L E ++E
Sbjct: 499 ERKRKRMLSNRESARRSRMRKQKQLEDLTDEVSRLQSANKKLAENIEAKEE 651
>TC226196 UP|Q8L5W2 (Q8L5W2) BZIP transcription factor ATB2, complete
Length = 1518
Score = 49.3 bits (116), Expect = 2e-06
Identities = 27/77 (35%), Positives = 48/77 (62%)
Frame = +2
Query: 215 LDAAISPSSLMGTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYT 274
++ A S + G+LS Q G + ++ + +R++KRMI NRESA RSR R+Q +
Sbjct: 731 INMACSSGTSSGSLSLLQNSGSEEDLQAMMED---QRKRKRMISNRESARRSRMRKQKHL 901
Query: 275 QELELKVSRLEEENERL 291
+L +V++L +EN+++
Sbjct: 902 DDLVSQVAQLRKENQQI 952
>TC216016 similar to UP|P93839 (P93839) G/HBF-1, partial (61%)
Length = 1764
Score = 48.1 bits (113), Expect = 5e-06
Identities = 23/44 (52%), Positives = 32/44 (72%)
Frame = +3
Query: 251 RRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQ 294
+R +RM+ NRESA RSR R+QA+ ELE +VS+L EN L ++
Sbjct: 735 KRVRRMLSNRESARRSRRRKQAHLTELETQVSQLRSENSSLLKR 866
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.310 0.127 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,975,987
Number of Sequences: 63676
Number of extensions: 129112
Number of successful extensions: 1109
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 1064
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1090
length of query: 321
length of database: 12,639,632
effective HSP length: 97
effective length of query: 224
effective length of database: 6,463,060
effective search space: 1447725440
effective search space used: 1447725440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146910.4


