

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146866.3 + phase: 0
(482 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
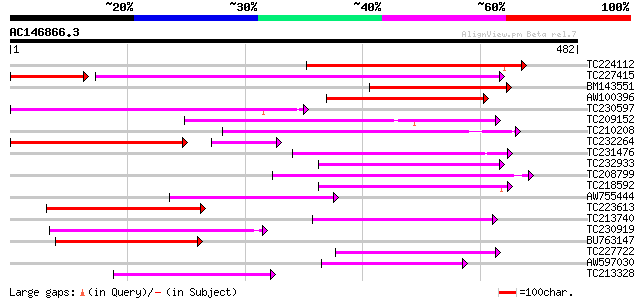
Score E
Sequences producing significant alignments: (bits) Value
TC224112 239 3e-63
TC227415 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (89%) 177 3e-55
BM143551 175 5e-44
AW100396 166 3e-41
TC230597 weakly similar to GB|AAP31960.1|30387589|BT006616 At1g1... 159 2e-39
TC209152 weakly similar to PIR|B96764|B96764 protein integral me... 145 3e-35
TC210208 139 2e-33
TC232264 weakly similar to UP|Q941E1 (Q941E1) At1g66760/F4N21_11... 106 2e-29
TC231476 weakly similar to GB|AAP31960.1|30387589|BT006616 At1g1... 102 3e-22
TC232933 similar to GB|AAN73299.1|25141209|BT002302 At3g26590/MF... 99 4e-21
TC208799 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, pa... 97 2e-20
TC218592 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, pa... 96 3e-20
AW755444 91 1e-18
TC223613 weakly similar to UP|Q9FNC1 (Q9FNC1) Emb|CAB89401.1, pa... 91 1e-18
TC213740 weakly similar to UP|TT12_ARATH (Q9LYT3) TRANSPARENT TE... 89 6e-18
TC230919 84 1e-16
BU763147 weakly similar to GP|15810018|gb AT5g38030/F16F17_30 {A... 82 4e-16
TC227722 73 3e-13
AW597030 similar to SP|Q9LYT3|TT12 TRANSPARENT TESTA 12 protein.... 73 3e-13
TC213328 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (27%) 72 4e-13
>TC224112
Length = 708
Score = 239 bits (609), Expect = 3e-63
Identities = 116/193 (60%), Positives = 144/193 (74%), Gaps = 6/193 (3%)
Frame = +1
Query: 253 VFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFT 312
+FPSSLS VSTR+GN+LGA +P KAR+S IV L + + G+ A++F ++RN W FT
Sbjct: 1 IFPSSLSFSVSTRVGNKLGAQKPSKARLSAIVGLSCSFMSGVLALVFALMVRNTWASMFT 180
Query: 313 NDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIF 372
D++I+ LTS+VLPI+GLCELGNCPQTTGCGVLRG+ARP +GANINLG FYLVGMPV+I+
Sbjct: 181 KDKDIITLTSMVLPIIGLCELGNCPQTTGCGVLRGTARPKVGANINLGCFYLVGMPVSIW 360
Query: 373 LGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELT------KSSTTSD 426
L F F GLW+GLLAAQGSCA+ MLVVLCRTDW + QRAK+LT S
Sbjct: 361 LAFFTGYDFQGLWLGLLAAQGSCAVTMLVVLCRTDWEFEAQRAKKLTGMGGAASGVDQSR 540
Query: 427 DVDAKLPIFMEGN 439
+VD + P+ E N
Sbjct: 541 EVDPEKPLKHESN 579
>TC227415 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (89%)
Length = 1748
Score = 177 bits (448), Expect(2) = 3e-55
Identities = 96/348 (27%), Positives = 182/348 (51%), Gaps = 1/348 (0%)
Frame = +2
Query: 74 ISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRIYLRTQGITLPLTY 133
++ I++ + +L+F G+ I+S A F+ L+P +F + P++ +L+ Q I P Y
Sbjct: 548 LTVIYVFSEPMLIFLGESPRIASAAALFVYGLIPQIFAYAANFPIQKFLQAQSIVAPSAY 727
Query: 134 CSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYFSSVYKKSWISPS 193
SA ++++H+ ++++ V +G+ G S+ + L+ +VI ++ S +++W +
Sbjct: 728 ISAATLVVHLGMSWVAVYEIGLGLLGASLVLSLSWWIMVIGQYVYIVKSERCRRTWQGFT 907
Query: 194 LDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIASMGILIQTTSLVYV 253
+ G L+ + V +CLE W+++ ++++ GLL NP+ + S+ I + V++
Sbjct: 908 WEAFSGLYGFFKLSAASAVMLCLETWYFQILVLLAGLLPNPELALDSLSICTTISGWVFM 1087
Query: 254 FPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTN 313
+ S R+ NELGA P+ A S++V ++ ++ + A L +R+ FT
Sbjct: 1088ISVGFNAAASVRVSNELGARSPKSASFSVVVVTVISFIISVIAALVVLALRDVISYAFTG 1267
Query: 314 DREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFL 373
E+ S + P++ L + N Q GV G A +N+G +Y VG+P+ L
Sbjct: 1268GEEVAAAVSDLCPLLALSLVLNGIQPVLSGVAVGCGWQAFVAYVNVGCYYGVGIPLGAVL 1447
Query: 374 GFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQR-AKELTK 420
GF + G G+W+G+L +++L V RTDW +V+ AK LTK
Sbjct: 1448GFYFQFGAKGIWLGMLGGTVMQTIILLWVTFRTDWTKEVEEAAKRLTK 1591
Score = 57.0 bits (136), Expect(2) = 3e-55
Identities = 26/67 (38%), Positives = 44/67 (64%)
Frame = +1
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
M + IF G+LG +ELA SL + Y ++ G+ +E +CGQA+GA+++ +LG+ +
Sbjct: 328 MSTQIFSGHLGNLELAAASLGNTGIQMFAYGLMLGMGSAVETLCGQAFGAQKYGMLGVYM 507
Query: 61 QRTVLLL 67
QR+ +LL
Sbjct: 508 QRSTILL 528
>BM143551
Length = 428
Score = 175 bits (443), Expect = 5e-44
Identities = 84/120 (70%), Positives = 93/120 (77%)
Frame = +2
Query: 307 WGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVG 366
W FT D EI+ LTS+VLPI+GLCELGNCPQTT CGVLRG+ARP +GANINLG FYLVG
Sbjct: 8 WASMFTRDAEIIALTSMVLPIIGLCELGNCPQTTVCGVLRGTARPKLGANINLGCFYLVG 187
Query: 367 MPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTKSSTTSD 426
MPVA+ L F A F GLW+GLLAAQ SC ML+VL RT+W QVQRAKELT SS D
Sbjct: 188 MPVAVRLSFFAGFDFKGLWLGLLAAQASCMFTMLIVLARTNWEGQVQRAKELTSSSEEQD 367
>AW100396
Length = 426
Score = 166 bits (419), Expect = 3e-41
Identities = 83/139 (59%), Positives = 104/139 (74%), Gaps = 1/139 (0%)
Frame = -3
Query: 270 LGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVLPIVG 329
LGA +P KA S IV L + +LG+ A++FT L+R+ W FT D+EI+ LTS VLP++G
Sbjct: 424 LGAQKPSKAEFSSIVGLSCSFMLGVFALVFTILVRHIWANMFTQDKEIITLTSFVLPVIG 245
Query: 330 LCE-LGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGL 388
L + + + GCGVLRG+ARP +GANINLG FYLVGMPVA++LGF A F GLW+GL
Sbjct: 244 LLQPWEHVLKQPGCGVLRGTARPKVGANINLGCFYLVGMPVAVWLGFFAGFDFQGLWLGL 65
Query: 389 LAAQGSCAMLMLVVLCRTD 407
LAAQGSCA+ MLVVL RTD
Sbjct: 64 LAAQGSCAVTMLVVLSRTD 8
>TC230597 weakly similar to GB|AAP31960.1|30387589|BT006616 At1g15170
{Arabidopsis thaliana;} , partial (51%)
Length = 1061
Score = 159 bits (403), Expect = 2e-39
Identities = 92/305 (30%), Positives = 156/305 (50%), Gaps = 51/305 (16%)
Frame = +3
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
++S + +G+LGE+ L+ +L+I + +TG+S++ G+A G+E ICGQAYG +Q++ +G+
Sbjct: 144 VVSTMIVGHLGELYLSSAALAISLSGVTGFSLLMGMASGLETICGQAYGGQQYQRIGIQT 323
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
+ L+ SIP+S +WINM+ IL+F GQD IS A F ++LVP LF +IL PL
Sbjct: 324 YTAIFSLILVSIPVSLLWINMETILVFIGQDPLISHEAGKFTIWLVPALFAYAILQPLVR 503
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
Y + Q + LP+ S V++++H+PL + LV ++ G ++A+ ++ + VI L ++
Sbjct: 504 YFQVQSLLLPMFASSCVTLIIHVPLCWALVFKTRLSNVGGALAVSISIWSNVIFLGLYMR 683
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSV-------------------------- 214
+ S K+ S++ KG AIP+ V V
Sbjct: 684 YFSACAKTRAPISMELFKGMWEFFRFAIPSAVMVW*LFFLFNQYFTYSCYQITKSFLVQQ 863
Query: 215 -------------------------CLEWWWYEFMIMMCGLLVNPKATIASMGILIQTTS 249
LEWW YE ++++ GLL NP+ + + + S
Sbjct: 864 IFYNVLYCFSKTSI*HNFCIHVDDGSLEWWSYELLVLLSGLLPNPQLETSVLSVW-YVCS 1040
Query: 250 LVYVF 254
++ VF
Sbjct: 1041IILVF 1055
>TC209152 weakly similar to PIR|B96764|B96764 protein integral membrane
protein F25P22.12 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (41%)
Length = 1166
Score = 145 bits (367), Expect = 3e-35
Identities = 81/271 (29%), Positives = 144/271 (52%), Gaps = 2/271 (0%)
Frame = +1
Query: 149 LVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYFSSVYKKSWISPSLDCIKGWSSLLSLAI 208
LV +G G ++A+ ++ V+ L+ ++ +S +K+ I S + + L LAI
Sbjct: 13 LVFKLGLGHVGAALAIGVSYWLNVVWLAIYMIYSPACQKTKIVFSSNALLSIPEFLKLAI 192
Query: 209 PTCVSVCLEWWWYEFMIMMCGLLVNPKATIASMGILIQTTSLVYVFPSSLSLGVSTRIGN 268
P+ + C EWW +E + ++ G+L NP+ A + I + TT+L Y P ++ STR+ N
Sbjct: 193 PSGLMFCFEWWSFEVLTLLAGILPNPQLETAVLSICLNTTTLHYFIPYAVGASASTRVSN 372
Query: 269 ELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVLPIV 328
ELGA P+ A+ ++ V + L + R+ G ++ND+E+++ + + P+
Sbjct: 373 ELGAGNPKTAKGAVRVVVILGVAEAAIVSTVFISCRHVLGYAYSNDKEVIDYVAEMAPL- 549
Query: 329 GLCELGNCPQTTGC--GVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWI 386
LC G G+ RG IGA +NLG++YLVG+P+ + LGF +L GLW+
Sbjct: 550 -LCVSVTADSLIGALSGIARGGGFQEIGAYVNLGAYYLVGIPMGLLLGFHLQLRAKGLWM 726
Query: 387 GLLAAQGSCAMLMLVVLCRTDWNLQVQRAKE 417
G L+ + +++ +V DW + +A+E
Sbjct: 727 GTLSGSLTQVIILAIVTALIDWQKEATKARE 819
>TC210208
Length = 743
Score = 139 bits (351), Expect = 2e-33
Identities = 82/253 (32%), Positives = 136/253 (53%)
Frame = +2
Query: 182 SSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIASM 241
S +K++W S + + + LA+P+ VCLE+W +E ++ + GL+ + + T + +
Sbjct: 14 SKKFKQTWKGFSTHSFRYVFTNMRLALPSAAMVCLEYWAFEVLVFLAGLMPDSQITTSLI 193
Query: 242 GILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTT 301
I I T + Y+ LS STR+ NELGA P++A+ +M V+L L+++LGL +L
Sbjct: 194 AICINTEFIAYMITYGLSAAASTRVSNELGAGNPERAKHAMSVTLKLSLLLGLCFVLALG 373
Query: 302 LMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGS 361
N W +FF++ I + + V P++ + L + Q GV RG + A INL +
Sbjct: 374 FGHNIWIQFFSDSSTIKKEFASVTPLLAISILLDAIQGVLSGVSRGCGWQHLAAYINLAT 553
Query: 362 FYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTKS 421
FYL+G+P++ FLGF L + GLWIGL+ +LC++ R K TK
Sbjct: 554 FYLIGLPISCFLGFKTNLQYKGLWIGLICG----------LLCQSGTLFLFIRRKXWTKL 703
Query: 422 STTSDDVDAKLPI 434
+ D+ D + P+
Sbjct: 704 DLSRDN-DKERPL 739
>TC232264 weakly similar to UP|Q941E1 (Q941E1) At1g66760/F4N21_11, partial
(58%)
Length = 701
Score = 106 bits (265), Expect(2) = 2e-29
Identities = 50/152 (32%), Positives = 94/152 (60%), Gaps = 1/152 (0%)
Frame = +1
Query: 1 MISMIFLGYLGEM-ELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLT 59
++S++ +G+ G + +G +++ FA +TG+SV+ G++ +E +CGQ YGA++++ G
Sbjct: 58 VVSLMMVGHFGILVSFSGVAIATSFAEVTGFSVLLGMSGALETLCGQTYGAEEYRKFGNY 237
Query: 60 LQRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLR 119
++ L +PIS +WI +ILL QD EIS A+ + ++L+P LF ++L L
Sbjct: 238 TWCAIVTLTLVCLPISLVWIFTDKILLLFSQDPEISHAAREYCIYLIPALFGHAVLQALT 417
Query: 120 IYLRTQGITLPLTYCSAVSVLLHIPLNFLLVV 151
Y +TQ + P+ + S ++ LH+P+ + LV+
Sbjct: 418 RYFQTQSMIFPMVFSSITALCLHVPICWSLVL 513
Score = 40.8 bits (94), Expect(2) = 2e-29
Identities = 18/60 (30%), Positives = 34/60 (56%)
Frame = +2
Query: 172 VILLSSFLYFSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLL 231
V+ L+ ++ +S +K+ I S + + L LAIP+ + C EWW +E + ++ G+L
Sbjct: 521 VVWLAIYMIYSPACQKTKIVFSSNALLSIPEFLKLAIPSGLMFCFEWWSFEVLTLLAGIL 700
>TC231476 weakly similar to GB|AAP31960.1|30387589|BT006616 At1g15170
{Arabidopsis thaliana;} , partial (32%)
Length = 734
Score = 102 bits (255), Expect = 3e-22
Identities = 57/188 (30%), Positives = 101/188 (53%), Gaps = 1/188 (0%)
Frame = +2
Query: 241 MGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFT 300
+ I + T S ++ P ++ STRI NELGA P A ++++ ++ A++
Sbjct: 8 LSICLNTISTLFSIPFGIAAAASTRISNELGAGNPHAAHVAVLAAMSFAIMETAIVSGTL 187
Query: 301 TLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLG 360
+ R+ +G F+N++E+++ +++ P++ + + + Q GV RG IG +NLG
Sbjct: 188 FVCRHDFGYIFSNEKEVVDYVTVMAPLICISVILDSIQGVLAGVARGCGWQHIGVYVNLG 367
Query: 361 SFYLVGMPVAIFLGFVAKLGFPGLWIGL-LAAQGSCAMLMLVVLCRTDWNLQVQRAKELT 419
+FYL G+PVA L F+AK+ GLWIG+ + A C + + C +W Q +A++
Sbjct: 368 AFYLCGIPVAATLAFLAKMRGKGLWIGVQVGAFVQCILFSTITSC-INWEQQAIKARKRL 544
Query: 420 KSSTTSDD 427
S S D
Sbjct: 545 FDSEISAD 568
>TC232933 similar to GB|AAN73299.1|25141209|BT002302 At3g26590/MFE16_11
{Arabidopsis thaliana;} , partial (30%)
Length = 545
Score = 99.0 bits (245), Expect = 4e-21
Identities = 47/158 (29%), Positives = 89/158 (55%)
Frame = +1
Query: 263 STRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTS 322
S R+ NELGA+ P+ A+ S++V++ + ++G+ + + RN + FF+ND E+ +
Sbjct: 55 SVRVSNELGASHPRTAKFSLLVAMITSTLIGVMLSMVLIIFRNHYPFFFSNDSEVRAMVV 234
Query: 323 IVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFP 382
+ P++ LC + N Q GV G+ + A +N+ +YL G+P+ + LG+ +G
Sbjct: 235 ELTPMLALCIVINNVQPVLSGVAVGAGWQALVAYVNIACYYLFGIPLGLVLGYKLDMGVM 414
Query: 383 GLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTK 420
G+W G+L+ ++ ++ RTDWN + A++ K
Sbjct: 415 GIWSGMLSGTILQTCVLFFLVYRTDWNKEASLAEDRIK 528
Score = 28.1 bits (61), Expect = 9.1
Identities = 42/164 (25%), Positives = 70/164 (42%), Gaps = 19/164 (11%)
Frame = +1
Query: 56 LGLTLQRTVLL-LLSTSIPISFIWINMKRILL--------FSGQDLEISSMAQSFILFLV 106
LG + RT LL I + I + + +L+ F D E+ +M ++ L
Sbjct: 76 LGASHPRTAKFSLLVAMITSTLIGVMLSMVLIIFRNHYPFFFSNDSEVRAM----VVELT 243
Query: 107 PDLFLLSILHPLRIYLRTQGITLPLTYCSAVSV-------LLHIPLNFLLVVHFQMGIAG 159
P L L +++ ++ L G+ + + + V+ L IPL +L MG+ G
Sbjct: 244 PMLALCIVINNVQPVL--SGVAVGAGWQALVAYVNIACYYLFGIPLGLVLGYKLDMGVMG 417
Query: 160 VSIAMVLTNLNLVILLSSFLYFSSVYKKSW---ISPSLDCIKGW 200
+ M L+ IL + L+F VY+ W S + D IK W
Sbjct: 418 IWSGM----LSGTILQTCVLFF-LVYRTDWNKEASLAEDRIKQW 534
>TC208799 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, partial (46%)
Length = 924
Score = 97.1 bits (240), Expect = 2e-20
Identities = 58/222 (26%), Positives = 107/222 (48%)
Frame = +2
Query: 224 MIMMCGLLVNPKATIASMGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMI 283
++++ G + N + I ++ I + + S R+ NELG + A+ S+I
Sbjct: 14 LVLLTGNMKNAEVEIDALSICLNINGWEMMISLGFMAAASVRVANELGRGSAKAAKFSII 193
Query: 284 VSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCG 343
VS+ ++ +G +F R + FT+++E+ + P++ + L N Q G
Sbjct: 194 VSVLTSLAIGFLLFIFFLFFRERLAYIFTSNKEVAFAVGDLSPLLSVSILLNSVQPVLSG 373
Query: 344 VLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVL 403
V G+ +I A +N+G +Y +G+PV I LG V L G+WIG+L ++++V+
Sbjct: 374 VAIGAGWQSIVAYVNMGCYYAIGIPVGIVLGNVLDLQVKGIWIGMLFGTLIQTIVLIVIT 553
Query: 404 CRTDWNLQVQRAKELTKSSTTSDDVDAKLPIFMEGNVNKNNV 445
+T+W+ QV A++ + D D E V + NV
Sbjct: 554 YKTNWDEQVTIAQKRISRWSKVDSPD------HENEVERKNV 661
>TC218592 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, partial (37%)
Length = 715
Score = 96.3 bits (238), Expect = 3e-20
Identities = 51/174 (29%), Positives = 93/174 (53%), Gaps = 9/174 (5%)
Frame = +3
Query: 263 STRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTS 322
S R+ NELG + A+ S++V++ ++ +G LF +R + FT+++++ +
Sbjct: 27 SVRVANELGKGSSKAAKFSIVVTVLTSLAIGFVLFLFFLFLRGKLAYIFTSNKDVADAVG 206
Query: 323 IVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFP 382
+ P++ + L N Q GV G+ +I A +N+G +Y++G+PV + LG V L
Sbjct: 207 DLSPLLAISILLNSVQPVLSGVAIGAGWQSIVAYVNIGCYYIIGIPVGVVLGNVLNLQVK 386
Query: 383 GLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAK---------ELTKSSTTSDD 427
G+WIG+L +++ V+ +TDW+ QV +A+ E + TSDD
Sbjct: 387 GIWIGMLFGTFIQTVVLTVITYKTDWDEQVTKARNRINKWSKVESDHETITSDD 548
>AW755444
Length = 432
Score = 90.9 bits (224), Expect = 1e-18
Identities = 47/143 (32%), Positives = 80/143 (55%)
Frame = +1
Query: 137 VSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYFSSVYKKSWISPSLDC 196
V ++LHIP+ ++LV +G G +I++ ++ V+LL + + +K+ I+ +
Sbjct: 1 VVLVLHIPICWVLVFGLGLGQNGAAISIGISYWLSVMLLLIYTKYYPSCQKTKIALGSNA 180
Query: 197 IKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIASMGILIQTTSLVYVFPS 256
++ LAIP+ + +C EWW +E ++++ GLL NPK + + I + +L Y P
Sbjct: 181 LRSIKEFFFLAIPSALMICFEWWSFELVVILAGLLPNPKLETSVLSICLNICTLHYFIPY 360
Query: 257 SLSLGVSTRIGNELGANRPQKAR 279
VSTR+ NEL A RPQ AR
Sbjct: 361 GTGAAVSTRVSNELXARRPQAAR 429
>TC223613 weakly similar to UP|Q9FNC1 (Q9FNC1) Emb|CAB89401.1, partial (15%)
Length = 460
Score = 90.5 bits (223), Expect = 1e-18
Identities = 41/135 (30%), Positives = 88/135 (64%)
Frame = +1
Query: 32 VISGLAMGMEPICGQAYGAKQWKILGLTLQRTVLLLLSTSIPISFIWINMKRILLFSGQD 91
++ G+A+ + +CGQAYGAK++ ++G+ LQR+ ++L ++I + ++I IL GQD
Sbjct: 7 ILLGMAIALSTLCGQAYGAKEYDMMGVYLQRSWIVLFLSAICLLPLFIFTSPILTLLGQD 186
Query: 92 LEISSMAQSFILFLVPDLFLLSILHPLRIYLRTQGITLPLTYCSAVSVLLHIPLNFLLVV 151
I+ +A++ ++ +P LF + + + +L++Q + ++Y +A+S+++H+ L++L +
Sbjct: 187 ESIAQVARTISIWSIPVLFAYIVSNSCQTFLQSQSKNVIISYLAALSIIIHVSLSWLFTM 366
Query: 152 HFQMGIAGVSIAMVL 166
F+ GI G I+ +L
Sbjct: 367 QFKYGIPGAMISTIL 411
>TC213740 weakly similar to UP|TT12_ARATH (Q9LYT3) TRANSPARENT TESTA 12
protein, partial (20%)
Length = 733
Score = 88.6 bits (218), Expect = 6e-18
Identities = 43/157 (27%), Positives = 86/157 (54%)
Frame = +3
Query: 258 LSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREI 317
++ +S R+ N LG + P+ A S V++F +++LG+ M ++++ K FT+ ++
Sbjct: 39 INTAISVRVSNTLGMSHPRAAIYSFCVTMFQSLLLGILFMTVIFFSKDEFAKIFTDSEDM 218
Query: 318 LELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVA 377
+ + + ++G+ + N GV GS + INL +Y+VG+P+ IFLGF
Sbjct: 219 ILAAADLAYLLGVTIVLNSASQVMSGVAIGSGWQVMVGYINLACYYIVGLPIGIFLGFKL 398
Query: 378 KLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQR 414
LG GLW G + +++ ++ +T+W+ +V++
Sbjct: 399 HLGVKGLWGGTMCGSILQTLVLFTIIWKTNWSKEVEQ 509
>TC230919
Length = 1034
Score = 84.3 bits (207), Expect = 1e-16
Identities = 50/185 (27%), Positives = 98/185 (52%)
Frame = +2
Query: 35 GLAMGMEPICGQAYGAKQWKILGLTLQRTVLLLLSTSIPISFIWINMKRILLFSGQDLEI 94
GL+ +E + Q +GA+++++LG+ LQ + ++ L SI IS IW + IL+ Q +I
Sbjct: 26 GLSGALETLXXQGFGAEEYQMLGIYLQASCIISLIFSIIISIIWFYTEPILVLLHQSQDI 205
Query: 95 SSMAQSFILFLVPDLFLLSILHPLRIYLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQ 154
+ + FL+P LF LS L + +L+TQ + L SA+ +L+HI + L+
Sbjct: 206 ARTTSLYTKFLIPGLFALSFLQNILRFLQTQSVVKSLVVFSAIPLLVHIFIA*ALIFCTD 385
Query: 155 MGIAGVSIAMVLTNLNLVILLSSFLYFSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSV 214
+ G +A+ ++ + LL ++ ++ ++++W S + + L LA+ +S
Sbjct: 386 LSFIGAPVAVSISLWISIPLLVMYIMYAERFRQTWTGFSFESFNYIFTDLKLAL---LSA 556
Query: 215 CLEWW 219
+ W+
Sbjct: 557 AMVWY 571
>BU763147 weakly similar to GP|15810018|gb AT5g38030/F16F17_30 {Arabidopsis
thaliana}, partial (46%)
Length = 429
Score = 82.4 bits (202), Expect = 4e-16
Identities = 40/125 (32%), Positives = 79/125 (63%)
Frame = -3
Query: 40 MEPICGQAYGAKQWKILGLTLQRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQ 99
+E +CGQA GA + +LG+ +QR+ +LLLST+ + ++I ++L GQD EIS A
Sbjct: 418 LETLCGQAVGAGKLDMLGVYMQRSWVLLLSTACVLCPLYIFAGQVLKLIGQDTEISEAAG 239
Query: 100 SFILFLVPDLFLLSILHPLRIYLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAG 159
+F ++++P LF ++ P+ +L+ Q + + + ++++LH L++LL+V + G+ G
Sbjct: 238 TFAIWMIPQLFAYALNFPVAKFLQAQSKVMVIAAIAGMAMVLHPVLSWLLMVKLEWGLVG 59
Query: 160 VSIAM 164
++ +
Sbjct: 58 AAVVL 44
>TC227722
Length = 676
Score = 72.8 bits (177), Expect = 3e-13
Identities = 40/140 (28%), Positives = 72/140 (50%)
Frame = +1
Query: 278 ARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCP 337
A+ S V +F ++ LG+ M R+ + FTN + + + + ++ + + N
Sbjct: 7 AKYSFYVIVFQSLFLGIFFMAIILATRDYYAIIFTNSEVLHKAVAKLGYLLAVTMVLNSV 186
Query: 338 QTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAM 397
Q GV G + A IN+G +YL G+P+ LG+ A LG GLW G++ +
Sbjct: 187 QPVVSGVAIGGGWQALVAYINIGCYYLFGLPLGFLLGYEANLGVEGLWGGMICGIVIQTL 366
Query: 398 LMLVVLCRTDWNLQVQRAKE 417
L+L++L +T+W +V++ E
Sbjct: 367 LLLLILYKTNWKKEVEQTTE 426
>AW597030 similar to SP|Q9LYT3|TT12 TRANSPARENT TESTA 12 protein. [Mouse-ear
cress] {Arabidopsis thaliana}, partial (25%)
Length = 401
Score = 72.8 bits (177), Expect = 3e-13
Identities = 38/124 (30%), Positives = 66/124 (52%)
Frame = +1
Query: 266 IGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVL 325
+ NELGA+ P+ A+ S+ V ++++ + + R K FT+D ++++ S +
Sbjct: 10 VSNELGASHPRVAKFSVFVVNGTSILISVVFCTIILIFRVSLSKLFTSDSDVIDAVSNLT 189
Query: 326 PIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLW 385
P++ + N Q GV GS + A +NL S+Y+VG+ V LGF LG G+W
Sbjct: 190 PLLAISVFFNGIQPILSGVAIGSGWQALVAYVNLASYYVVGLTVGCVLGFKTSLGVAGIW 369
Query: 386 IGLL 389
G++
Sbjct: 370 WGMI 381
>TC213328 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (27%)
Length = 423
Score = 72.4 bits (176), Expect = 4e-13
Identities = 34/138 (24%), Positives = 71/138 (50%)
Frame = +3
Query: 89 GQDLEISSMAQSFILFLVPDLFLLSILHPLRIYLRTQGITLPLTYCSAVSVLLHIPLNFL 148
G+ I+S A + L+P +F ++ P++ +L+ Q I P Y S ++L+H+ L++
Sbjct: 9 GESPRIASAAALLVYGLIPQIFAYAVNFPIQKFLQAQSIVAPSAYISTATLLVHLVLSYF 188
Query: 149 LVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYFSSVYKKSWISPSLDCIKGWSSLLSLAI 208
+V +G+ G S+ + ++ +VI ++ S K +W S G L+
Sbjct: 189 VVYEVGLGLLGASLVLSVSWWIIVIAQFVYIVKSEKCKHTWRGFSFRAFSGLPEFFKLSA 368
Query: 209 PTCVSVCLEWWWYEFMIM 226
+ V +CLE W+++ +++
Sbjct: 369 ASAVMLCLETWYFQILVL 422
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.141 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,904,525
Number of Sequences: 63676
Number of extensions: 390012
Number of successful extensions: 3086
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 3028
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3078
length of query: 482
length of database: 12,639,632
effective HSP length: 101
effective length of query: 381
effective length of database: 6,208,356
effective search space: 2365383636
effective search space used: 2365383636
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146866.3


