

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.9 + phase: 0
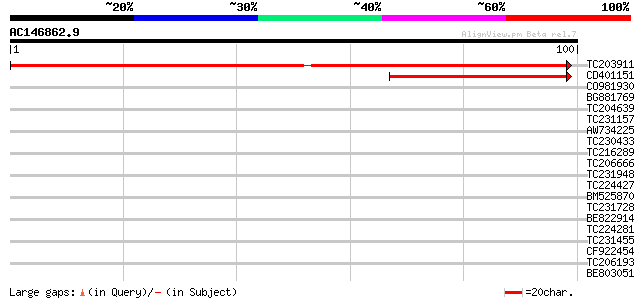
(100 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203911 similar to UP|Q95RQ4 (Q95RQ4) LD16074p (CG6393-PA), par... 193 1e-50
CD401151 similar to GP|23307683|gb| NADH dehydrogenase subunit 1... 76 2e-15
CO981930 30 0.25
BG881769 28 0.55
TC204639 UP|RSP4_SOYBN (O22518) 40S ribosomal protein SA (p40), ... 28 0.55
TC231157 similar to GB|CAC82909.1|25136916|ATH297948 cellulose s... 27 1.2
AW734225 27 1.2
TC230433 similar to UP|Q9LRH6 (Q9LRH6) ZIM, partial (45%) 27 1.6
TC216289 UP|Q8W3Y5 (Q8W3Y5) Flavonoid 3'-hydroxylase, complete 27 2.1
TC206666 27 2.1
TC231948 similar to UP|Q6NNI3 (Q6NNI3) At5g28490, partial (69%) 27 2.1
TC224427 27 2.1
BM525870 27 2.1
TC231728 26 2.7
BE822914 similar to GP|5882748|gb| Contains 2 PF|01535 DUF domai... 26 2.7
TC224281 26 2.7
TC231455 weakly similar to UP|Q9SPL8 (Q9SPL8) Dehydrin, partial ... 26 2.7
CF922454 26 2.7
TC206193 weakly similar to UP|Q9ZR88 (Q9ZR88) Bifunctional nucle... 26 2.7
BE803051 similar to PIR|T10863|T108 extensin precursor - kidney ... 26 3.6
>TC203911 similar to UP|Q95RQ4 (Q95RQ4) LD16074p (CG6393-PA), partial (12%)
Length = 2055
Score = 193 bits (491), Expect = 1e-50
Identities = 84/99 (84%), Positives = 92/99 (92%)
Frame = +1
Query: 1 MHRGFKFSITNSIMWQTADRSLIWKQSPEQPSAYPHFPFKARDMNTPIIQYHHPCCYCGV 60
MHRGFKFSITNSIMWQTADR L+WKQSPEQP +PH P+KA+D+ TP++QYH P C+CGV
Sbjct: 1075 MHRGFKFSITNSIMWQTADRPLMWKQSPEQPIVFPHCPYKAKDITTPVVQYH-PFCFCGV 1251
Query: 61 KSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWAS 99
KSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWAS
Sbjct: 1252 KSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWAS 1368
>CD401151 similar to GP|23307683|gb| NADH dehydrogenase subunit 1 {Brugia
malayi}, partial (8%)
Length = 532
Score = 76.3 bits (186), Expect = 2e-15
Identities = 31/32 (96%), Positives = 31/32 (96%)
Frame = -2
Query: 68 RKPGPKQGSLFFGCGNWTATRGARCHYFEWAS 99
RKPGPKQGSLF GCGNWTATRGARCHYFEWAS
Sbjct: 531 RKPGPKQGSLFLGCGNWTATRGARCHYFEWAS 436
>CO981930
Length = 724
Score = 29.6 bits (65), Expect = 0.25
Identities = 14/38 (36%), Positives = 20/38 (51%), Gaps = 3/38 (7%)
Frame = -3
Query: 66 MVRKPGPKQGSLFFGCGNWTATRG---ARCHYFEWASA 100
+V+K GP G F+ C + A C YF+WAS+
Sbjct: 287 VVKKQGPNFGRRFYVCAHAEGPASNLEANCGYFKWASS 174
>BG881769
Length = 399
Score = 28.5 bits (62), Expect = 0.55
Identities = 16/42 (38%), Positives = 22/42 (52%)
Frame = -3
Query: 1 MHRGFKFSITNSIMWQTADRSLIWKQSPEQPSAYPHFPFKAR 42
+H+ ITNS+MW+ R L+ +QS YP P K R
Sbjct: 169 LHKSS*QKITNSMMWRLDPRQLLERQSLR---VYPIIPSKFR 53
>TC204639 UP|RSP4_SOYBN (O22518) 40S ribosomal protein SA (p40), complete
Length = 1244
Score = 28.5 bits (62), Expect = 0.55
Identities = 16/35 (45%), Positives = 20/35 (56%)
Frame = -1
Query: 21 SLIWKQSPEQPSAYPHFPFKARDMNTPIIQYHHPC 55
SL+W EQ + PF+AR TP +Q HHPC
Sbjct: 725 SLLWFSIEEQINH--DIPFEARANGTPHLQ-HHPC 630
>TC231157 similar to GB|CAC82909.1|25136916|ATH297948 cellulose synthase-like
protein {Arabidopsis thaliana;} , partial (23%)
Length = 932
Score = 27.3 bits (59), Expect = 1.2
Identities = 10/29 (34%), Positives = 15/29 (51%)
Frame = -1
Query: 27 SPEQPSAYPHFPFKARDMNTPIIQYHHPC 55
S S F + DM+TP + +H+PC
Sbjct: 482 SSSPSSGVADFEVRVNDMSTPAMTFHNPC 396
>AW734225
Length = 412
Score = 27.3 bits (59), Expect = 1.2
Identities = 15/45 (33%), Positives = 21/45 (46%), Gaps = 6/45 (13%)
Frame = +1
Query: 16 QTADRSLIWKQSPEQPSAYP------HFPFKARDMNTPIIQYHHP 54
++ DRSL+ + P Q P HFP TP I +H+P
Sbjct: 121 RSLDRSLLVRSHPPQVPIPPPLRCPRHFPLSRPRARTPSIGWHNP 255
>TC230433 similar to UP|Q9LRH6 (Q9LRH6) ZIM, partial (45%)
Length = 889
Score = 26.9 bits (58), Expect = 1.6
Identities = 14/37 (37%), Positives = 19/37 (50%), Gaps = 1/37 (2%)
Frame = +1
Query: 55 CCYCGVKSSRGMVRKPGPK-QGSLFFGCGNWTATRGA 90
C +CG+ S + + GP SL CG + A RGA
Sbjct: 370 CTHCGISSKSTPMMRRGPSGPRSLCNACGLFWANRGA 480
>TC216289 UP|Q8W3Y5 (Q8W3Y5) Flavonoid 3'-hydroxylase, complete
Length = 1883
Score = 26.6 bits (57), Expect = 2.1
Identities = 11/28 (39%), Positives = 12/28 (42%)
Frame = -1
Query: 31 PSAYPHFPFKARDMNTPIIQYHHPCCYC 58
P Y PF + I YHH C YC
Sbjct: 1766 PKTYEMTPFHVFGSSHTNILYHHVCFYC 1683
>TC206666
Length = 1493
Score = 26.6 bits (57), Expect = 2.1
Identities = 9/18 (50%), Positives = 10/18 (55%)
Frame = +1
Query: 22 LIWKQSPEQPSAYPHFPF 39
L+WK SP P HF F
Sbjct: 76 LLWKHSPSSPKTLAHFSF 129
>TC231948 similar to UP|Q6NNI3 (Q6NNI3) At5g28490, partial (69%)
Length = 1023
Score = 26.6 bits (57), Expect = 2.1
Identities = 11/20 (55%), Positives = 11/20 (55%)
Frame = -1
Query: 51 YHHPCCYCGVKSSRGMVRKP 70
YH CC C K S GM R P
Sbjct: 447 YHFHCCCCLNKLSSGMPRSP 388
>TC224427
Length = 794
Score = 26.6 bits (57), Expect = 2.1
Identities = 11/31 (35%), Positives = 16/31 (51%)
Frame = +1
Query: 27 SPEQPSAYPHFPFKARDMNTPIIQYHHPCCY 57
S +P+A+ F A + P + HHP CY
Sbjct: 280 SANRPAAFSVFSKSAESPSPPNPKRHHPVCY 372
>BM525870
Length = 436
Score = 26.6 bits (57), Expect = 2.1
Identities = 17/48 (35%), Positives = 23/48 (47%)
Frame = +1
Query: 29 EQPSAYPHFPFKARDMNTPIIQYHHPCCYCGVKSSRGMVRKPGPKQGS 76
E P P P + T QYHH CC G ++S G+ R+ P + S
Sbjct: 46 ETPLGSPLLP----NPKTTSYQYHHLCCR-GSRNSSGV*RQRNPFERS 174
>TC231728
Length = 470
Score = 26.2 bits (56), Expect = 2.7
Identities = 7/12 (58%), Positives = 10/12 (83%)
Frame = +1
Query: 46 TPIIQYHHPCCY 57
TP+++ HPCCY
Sbjct: 43 TPVLEPQHPCCY 78
>BE822914 similar to GP|5882748|gb| Contains 2 PF|01535 DUF domains.
{Arabidopsis thaliana}, partial (17%)
Length = 610
Score = 26.2 bits (56), Expect = 2.7
Identities = 13/50 (26%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Frame = +2
Query: 24 WKQSPEQPSAYPHFPFKARDMNTPIIQYHHPCCYCGVKSSR-GMVRKPGP 72
W++ P+ +P FPF A+ N + + C C + ++R+P P
Sbjct: 191 WQRGSPHPTKHPEFPFSAK--NGKLNVCNRSCTACLTREDPVTLLRRPHP 334
>TC224281
Length = 660
Score = 26.2 bits (56), Expect = 2.7
Identities = 18/51 (35%), Positives = 22/51 (42%), Gaps = 2/51 (3%)
Frame = +2
Query: 33 AYPHFPFKARDMNTPII--QYHHPCCYCGVKSSRGMVRKPGPKQGSLFFGC 81
A P P R + +PI H C C SS ++R PGP SL C
Sbjct: 329 AVPPVPKLTRSITSPIWARSSRHWACECCGASSLHVLRHPGPHTLSLNVCC 481
>TC231455 weakly similar to UP|Q9SPL8 (Q9SPL8) Dehydrin, partial (7%)
Length = 718
Score = 26.2 bits (56), Expect = 2.7
Identities = 8/12 (66%), Positives = 9/12 (74%)
Frame = -1
Query: 45 NTPIIQYHHPCC 56
N P+I YH PCC
Sbjct: 115 NNPVIPYHVPCC 80
>CF922454
Length = 785
Score = 26.2 bits (56), Expect = 2.7
Identities = 12/31 (38%), Positives = 15/31 (47%)
Frame = -1
Query: 27 SPEQPSAYPHFPFKARDMNTPIIQYHHPCCY 57
SP PS P P +P + YH+PC Y
Sbjct: 305 SPSSPSPSPSSP-------SPSLPYHYPCHY 234
>TC206193 weakly similar to UP|Q9ZR88 (Q9ZR88) Bifunctional nuclease
(Fragment), partial (84%)
Length = 1973
Score = 26.2 bits (56), Expect = 2.7
Identities = 10/24 (41%), Positives = 14/24 (57%), Gaps = 2/24 (8%)
Frame = -1
Query: 52 HHPCCYCGVKSSRGMVR--KPGPK 73
H+PCC+C + G +R K PK
Sbjct: 1001 HNPCCFCNCRGPLGYLRSAKQRPK 930
>BE803051 similar to PIR|T10863|T108 extensin precursor - kidney bean,
partial (10%)
Length = 442
Score = 25.8 bits (55), Expect = 3.6
Identities = 12/41 (29%), Positives = 18/41 (43%)
Frame = +2
Query: 16 QTADRSLIWKQSPEQPSAYPHFPFKARDMNTPIIQYHHPCC 56
Q RSL+ Q P++ Y + P +P Y +P C
Sbjct: 29 QNTTRSLVLHQDPKKHYKYQNLPPSNYKNKSPSSPYKYPSC 151
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.137 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,614,993
Number of Sequences: 63676
Number of extensions: 127956
Number of successful extensions: 1106
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 1101
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1105
length of query: 100
length of database: 12,639,632
effective HSP length: 76
effective length of query: 24
effective length of database: 7,800,256
effective search space: 187206144
effective search space used: 187206144
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC146862.9


