

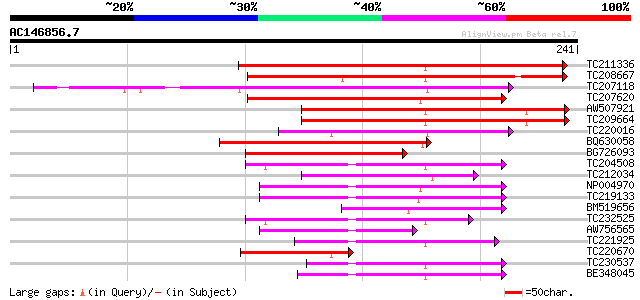
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146856.7 + phase: 0
(241 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211336 weakly similar to UP|Q8GVD9 (Q8GVD9) Cyclin D3, partial... 171 2e-43
TC208667 weakly similar to UP|Q84V88 (Q84V88) Cyclin D, partial ... 120 6e-28
TC207118 homologue to UP|Q6T2Z6 (Q6T2Z6) Cyclin d3, complete 116 1e-26
TC207620 similar to UP|Q8LK74 (Q8LK74) Cyclin D3.1 protein, part... 114 4e-26
AW507921 94 7e-20
TC209664 UP|Q6T2Z7 (Q6T2Z7) Cyclin d2, partial (69%) 93 1e-19
TC220016 similar to UP|Q7XAB6 (Q7XAB6) Cyclin D3-1, partial (51%) 90 1e-18
BQ630058 weakly similar to GP|3608179|dbj| cyclin D {Pisum sativ... 88 4e-18
BG726093 similar to GP|4160298|emb cyclin D2.1 protein {Nicotian... 76 2e-14
TC204508 homologue to UP|Q39855 (Q39855) Cyclin, partial (98%) 70 9e-13
TC212034 similar to UP|Q7XAB6 (Q7XAB6) Cyclin D3-1, partial (9%) 62 2e-10
NP004970 mitotic cyclin a1-type 61 4e-10
TC219133 UP|Q39879 (Q39879) Mitotic cyclin a2-type, complete 60 1e-09
BM519656 55 2e-08
TC232525 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type,... 54 5e-08
AW756565 similar to PIR|T07675|T07 cyclin a2-type mitosis-speci... 54 8e-08
TC221925 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type,... 51 5e-07
TC220670 similar to UP|Q8GZU3 (Q8GZU3) Cyclin D, partial (14%) 50 7e-07
TC230537 mitotic cyclin a2-type 49 2e-06
BE348045 similar to PIR|T07672|T07 cyclin a2-type mitosis-speci... 49 2e-06
>TC211336 weakly similar to UP|Q8GVD9 (Q8GVD9) Cyclin D3, partial (28%)
Length = 1351
Score = 171 bits (434), Expect = 2e-43
Identities = 84/141 (59%), Positives = 105/141 (73%), Gaps = 1/141 (0%)
Frame = +2
Query: 98 WLRNARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLS 157
W++ AR+ AI+WI T+A GF +TAYLS+ YFDRFLS+RSID K WAI+LLS+ACLS
Sbjct: 50 WVKRARVEAINWILKTRATLGFRFETAYLSVTYFDRFLSRRSIDSEKSWAIRLLSIACLS 229
Query: 158 IAAKMEEQSVPPLSEYPI-EYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKFC 216
+AAKMEE +VP LSE+ + +Y FE KVI+ MELL+LSTLEW+MG+ TPF +L YF TKFC
Sbjct: 230 LAAKMEECNVPGLSEFKLDDYSFEGKVIQKMELLVLSTLEWEMGIITPFDFLSYFITKFC 409
Query: 217 NGSRSETIITKATQHIVTMVK 237
S I K Q I T +K
Sbjct: 410 KESPPSPIFYKTMQLIFTTMK 472
>TC208667 weakly similar to UP|Q84V88 (Q84V88) Cyclin D, partial (24%)
Length = 1149
Score = 120 bits (301), Expect = 6e-28
Identities = 67/140 (47%), Positives = 92/140 (64%), Gaps = 4/140 (2%)
Frame = +3
Query: 102 ARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSI-DESKPWAIQLLSVACLSIAA 160
ARL A++WI A + F+ TA+LS+NYFDRFLS+ S+ +S WA QLLSVACLS+AA
Sbjct: 264 ARLDAVNWILKVHAYYEFSPVTAFLSVNYFDRFLSRCSLPQQSGGWAFQLLSVACLSLAA 443
Query: 161 KMEEQSVPPLSEYPI---EYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKFCN 217
KMEE VP L + + ++ F+ K + MEL ++S L+W++ TPF YLHYFFTK
Sbjct: 444 KMEESHVPFLLDLQLFQPKFVFQPKTDQRMELWVMSNLKWRLRSVTPFDYLHYFFTKL-- 617
Query: 218 GSRSETIITKATQHIVTMVK 237
S S IT A+ I++ +
Sbjct: 618 PSSSSQSITTASNLILSTTR 677
>TC207118 homologue to UP|Q6T2Z6 (Q6T2Z6) Cyclin d3, complete
Length = 1763
Score = 116 bits (290), Expect = 1e-26
Identities = 73/222 (32%), Positives = 113/222 (50%), Gaps = 18/222 (8%)
Frame = +3
Query: 11 LSSLMCHEQDESTLIFEEDEDESTFFINSSLDNNNNN---PWFLLDD----EEEYIQYLF 63
L +L C E +E++E+E + S + N P +L+ E+E + LF
Sbjct: 261 LDALYCDEGK-----WEDEEEEEEXYEESEVTTNTGTSLFPLLMLEQDLFWEDEELNSLF 425
Query: 64 KQETGLGFGSITHFLCYDDHDVEVEDDDSSKSL--------FWLRNARLHAIDWIFNTQA 115
+E + H YD +++ +D+ + S L R A++WI A
Sbjct: 426 SKE------KVQHEEAYDYNNLNSDDNSNDHSNNNNNVLSDSCLSQPRREAVEWILKVNA 587
Query: 116 KFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI 175
+GF+ TA L++ Y DRFL KPW IQL++V C+S+AAK+EE VP L + +
Sbjct: 588 HYGFSALTATLAVTYLDRFLLSFHFQREKPWMIQLVAVTCISLAAKVEETQVPLLLDLQV 767
Query: 176 E---YRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTK 214
+ Y FE K I+ MELL+LSTL+WKM TP ++L + +
Sbjct: 768 QDTKYVFEAKTIQRMELLVLSTLKWKMHPVTPLSFLDHIIRR 893
>TC207620 similar to UP|Q8LK74 (Q8LK74) Cyclin D3.1 protein, partial (51%)
Length = 1405
Score = 114 bits (285), Expect = 4e-26
Identities = 57/114 (50%), Positives = 72/114 (63%), Gaps = 4/114 (3%)
Frame = +3
Query: 102 ARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAK 161
AR A++W+ + + F+ TA LS+NYFDRFL KPW +QL +VACLSIAAK
Sbjct: 276 ARREAVEWMLKVNSHYSFSALTAVLSVNYFDRFLFSFRFQNDKPWMVQLAAVACLSIAAK 455
Query: 162 MEEQSVPPLSEY----PIEYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYF 211
+EE VP L + Y FE K IK ME+L+LSTL WKM PTP ++L YF
Sbjct: 456 VEETHVPFLIDLQQVDESRYLFEAKTIKKMEILVLSTLGWKMNPPTPLSFLDYF 617
>AW507921
Length = 445
Score = 93.6 bits (231), Expect = 7e-20
Identities = 51/118 (43%), Positives = 74/118 (62%), Gaps = 4/118 (3%)
Frame = +2
Query: 125 YLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI---EYRFEN 181
YLSINY DRFL + + + W +QLL+VACLS+AAK++E VP + + ++ FE
Sbjct: 5 YLSINYLDRFLFAYELPKGRVWTMQLLAVACLSLAAKLDETEVPLSLDLQVGESKFLFEA 184
Query: 182 KVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKFCNG-SRSETIITKATQHIVTMVKG 238
K I+ MELL+LSTL+W+M TPF +L YF K + S + I ++ Q I + +G
Sbjct: 185 KTIQRMELLVLSTLKWRMQAITPFTFLDYFLCKINDDQSPLRSSIMRSIQLISSTARG 358
>TC209664 UP|Q6T2Z7 (Q6T2Z7) Cyclin d2, partial (69%)
Length = 903
Score = 92.8 bits (229), Expect = 1e-19
Identities = 50/118 (42%), Positives = 74/118 (62%), Gaps = 4/118 (3%)
Frame = +2
Query: 125 YLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI---EYRFEN 181
YLSINY DRFL + + + W +QLL+VAC+S+AAK++E VP + + ++ FE
Sbjct: 8 YLSINYLDRFLCAYELPKGRVWTMQLLAVACVSLAAKLDETEVPLSLDLQVGESKFLFEA 187
Query: 182 KVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKFCNG-SRSETIITKATQHIVTMVKG 238
K I+ MELL+LSTL+W+M TPF +L YF K + S + I ++ Q I + +G
Sbjct: 188 KTIQRMELLVLSTLKWRMQAITPFTFLDYFLCKINDDQSPLRSSIMRSIQLISSTARG 361
>TC220016 similar to UP|Q7XAB6 (Q7XAB6) Cyclin D3-1, partial (51%)
Length = 921
Score = 89.7 bits (221), Expect = 1e-18
Identities = 48/111 (43%), Positives = 66/111 (59%), Gaps = 11/111 (9%)
Frame = +1
Query: 115 AKFGFTVQTAYLSINYFDRFL--------SKRSIDESKPWAIQLLSVACLSIAAKMEEQS 166
A++ F+ TA L++NY DRFL + + + + PW QL +VACLS+AAK EE
Sbjct: 13 ARYSFSTLTAVLAVNYLDRFLFSFRFQNDNNDNNNNNNPWLTQLSAVACLSLAAKFEETH 192
Query: 167 VPPLSEYPIE---YRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTK 214
VP + +E Y FE K +K ME+L+LSTL WKM TP ++L Y K
Sbjct: 193 VPLFIDLQVEESKYLFEAKTVKRMEILVLSTLGWKMNPVTPLSFLDYITRK 345
>BQ630058 weakly similar to GP|3608179|dbj| cyclin D {Pisum sativum}, partial
(19%)
Length = 433
Score = 87.8 bits (216), Expect = 4e-18
Identities = 45/91 (49%), Positives = 58/91 (63%), Gaps = 1/91 (1%)
Frame = +2
Query: 90 DDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQ 149
D++ WL NAR AI+WI A + F +TAYLS++YF+RFL + + K W +Q
Sbjct: 158 DNNFAKKTWLINAREEAINWILKVHAYYSFKPETAYLSVDYFNRFLLSHTFTQDKAWPLQ 337
Query: 150 LLSVACLSIAAKMEEQSVPPLSEYP-IEYRF 179
LLSV CLS+AAKMEE VP L + IE RF
Sbjct: 338 LLSVTCLSLAAKMEESKVPLLLDLQVIESRF 430
>BG726093 similar to GP|4160298|emb cyclin D2.1 protein {Nicotiana tabacum},
partial (29%)
Length = 412
Score = 75.9 bits (185), Expect = 2e-14
Identities = 37/69 (53%), Positives = 46/69 (66%)
Frame = +3
Query: 101 NARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAA 160
+ R A+DWI+ A F F + LS+NY DRFLS + K W++QLL+VACLSIAA
Sbjct: 12 SVRKEALDWIWKAHAYFDFGPCSLCLSVNYLDRFLSVYELPRGKSWSMQLLAVACLSIAA 191
Query: 161 KMEEQSVPP 169
KMEE VPP
Sbjct: 192 KMEEIKVPP 218
Score = 32.7 bits (73), Expect = 0.15
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = +1
Query: 176 EYRFENKVIKNMELLILSTLEWK 198
++ FE K I+ MELL+LSTL WK
Sbjct: 343 KFAFEAKDIQRMELLVLSTLRWK 411
>TC204508 homologue to UP|Q39855 (Q39855) Cyclin, partial (98%)
Length = 1283
Score = 70.1 bits (170), Expect = 9e-13
Identities = 38/115 (33%), Positives = 69/115 (59%), Gaps = 4/115 (3%)
Frame = +2
Query: 101 NARLHAI--DWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSI 158
N R+ AI DW+ + KF +++T YL+IN DRFL+ +++ + +QL+ ++ + +
Sbjct: 407 NERMRAILVDWLIDVHTKFELSLETLYLTINIIDRFLAVKTVPRRE---LQLVGISAMLM 577
Query: 159 AAKMEEQSVPPLSEYPI--EYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYF 211
A+K EE P ++++ + + ++ I ME IL+ LEW + +PTPF +L F
Sbjct: 578 ASKYEEIWPPEVNDFVCLSDRAYTHEQILAMEKTILNKLEWTLTVPTPFVFLVRF 742
>TC212034 similar to UP|Q7XAB6 (Q7XAB6) Cyclin D3-1, partial (9%)
Length = 698
Score = 62.4 bits (150), Expect = 2e-10
Identities = 32/81 (39%), Positives = 48/81 (58%), Gaps = 6/81 (7%)
Frame = +1
Query: 125 YLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPIEYR------ 178
YL+INY DRFL+ + I + KPWA +LL+V+C S+AAKM + +
Sbjct: 448 YLAINYLDRFLANQGILQPKPWANKLLAVSCFSLAAKMLKTEYSATDVQVLMNHGDGGAI 627
Query: 179 FENKVIKNMELLILSTLEWKM 199
FE + I+ ME ++L L+W+M
Sbjct: 628 FETQTIQRMEGIVLGALQWRM 690
>NP004970 mitotic cyclin a1-type
Length = 1047
Score = 61.2 bits (147), Expect = 4e-10
Identities = 34/107 (31%), Positives = 59/107 (54%), Gaps = 2/107 (1%)
Frame = +1
Query: 107 IDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQS 166
+DW+ ++ T +LS++Y DRFLS + +S+ +QLL V+ + IAAK EE
Sbjct: 358 VDWLVEVAEEYKLLSDTLHLSVSYIDRFLSVNPVTKSR---LQLLGVSSMLIAAKYEETD 528
Query: 167 VPPLSEY--PIEYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYF 211
P + E+ + ++ + ME IL +L+++MG PT +L +
Sbjct: 529 PPSVDEFCSITDNTYDKAEVVKMEADILKSLKFEMGNPTVSTFLRRY 669
>TC219133 UP|Q39879 (Q39879) Mitotic cyclin a2-type, complete
Length = 1804
Score = 59.7 bits (143), Expect = 1e-09
Identities = 34/107 (31%), Positives = 57/107 (52%), Gaps = 2/107 (1%)
Frame = +2
Query: 107 IDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQS 166
IDW+ ++ T YL++NY DR+LS ++ + +QLL VA + IA+K EE
Sbjct: 812 IDWLVEVAEEYRLVPDTLYLTVNYIDRYLSGNVMNRQR---LQLLGVASMMIASKYEEIC 982
Query: 167 VPPLSE--YPIEYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYF 211
P + E Y + + + + ME +L+ L+++M PT +L F
Sbjct: 983 APQVEEFCYITDNTYFKEEVLQMESAVLNFLKFEMTAPTVKCFLRRF 1123
>BM519656
Length = 428
Score = 55.5 bits (132), Expect = 2e-08
Identities = 27/76 (35%), Positives = 45/76 (58%), Gaps = 6/76 (7%)
Frame = +2
Query: 142 ESKPWAIQLLSVACLSIAAKMEEQSVP------PLSEYPIEYRFENKVIKNMELLILSTL 195
+ KPW ++L++V+C+S+A KM P L++ FE + I+ ME LIL L
Sbjct: 2 QPKPWVLRLIAVSCISLAVKMMRTEYPFTDVQALLNQSDGGIIFETQTIQRMEALILGAL 181
Query: 196 EWKMGLPTPFAYLHYF 211
+W+M TPF+++ +F
Sbjct: 182 QWRMRSITPFSFVAFF 229
>TC232525 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type, partial
(73%)
Length = 1242
Score = 54.3 bits (129), Expect = 5e-08
Identities = 31/101 (30%), Positives = 57/101 (55%), Gaps = 4/101 (3%)
Frame = +1
Query: 101 NARLHAI--DWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSI 158
NA++ +I DW+ KF +T YL++N DRFLS +++ + +QL+ ++ + I
Sbjct: 898 NAKMRSILVDWLIEVHRKFELMPETLYLTLNIVDRFLSVKAVPRRE---LQLVGISSMLI 1068
Query: 159 AAKMEEQSVPPLSEYPI--EYRFENKVIKNMELLILSTLEW 197
A+K EE P ++++ + + ++ + ME IL LEW
Sbjct: 1069ASKYEEIWAPEVNDFECISDNAYVSQQVLMMEKKILRKLEW 1191
>AW756565 similar to PIR|T07675|T07 cyclin a2-type mitosis-specific -
soybean, partial (28%)
Length = 472
Score = 53.5 bits (127), Expect = 8e-08
Identities = 25/67 (37%), Positives = 40/67 (59%)
Frame = +2
Query: 107 IDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQS 166
+DW+ ++ +T YL++NY DR LS +++ + IQLL V+C+ IA+K EE
Sbjct: 215 VDWLVEVAEEYRLVPETLYLTVNYLDRSLSWNAMNRQR---IQLLGVSCMMIASKYEEIC 385
Query: 167 VPPLSEY 173
P + EY
Sbjct: 386 APHVEEY 406
>TC221925 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type, partial
(46%)
Length = 802
Score = 50.8 bits (120), Expect = 5e-07
Identities = 27/89 (30%), Positives = 53/89 (59%), Gaps = 2/89 (2%)
Frame = +2
Query: 122 QTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI--EYRF 179
+T YL++ DRFLS +++ + +QL+ ++ + IA+K EE P ++++ + +
Sbjct: 5 ETLYLTLXIVDRFLSVKAVPRRE---LQLVGISSMLIASKYEEIWAPEVNDFECISDNAY 175
Query: 180 ENKVIKNMELLILSTLEWKMGLPTPFAYL 208
++ + ME IL LEW + +PTP+ +L
Sbjct: 176 VSQQVLMMEXXILRKLEWYLTVPTPYHFL 262
>TC220670 similar to UP|Q8GZU3 (Q8GZU3) Cyclin D, partial (14%)
Length = 521
Score = 50.4 bits (119), Expect = 7e-07
Identities = 23/49 (46%), Positives = 32/49 (64%), Gaps = 1/49 (2%)
Frame = +3
Query: 99 LRNARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFL-SKRSIDESKPW 146
L AR+ A++WI A + F+ TA L++NYFDRFL S R ++ KPW
Sbjct: 372 LEGARIEAVEWILKVNAHYSFSALTAVLAVNYFDRFLFSFRFQNDIKPW 518
>TC230537 mitotic cyclin a2-type
Length = 993
Score = 48.9 bits (115), Expect = 2e-06
Identities = 28/87 (32%), Positives = 47/87 (53%), Gaps = 2/87 (2%)
Frame = +1
Query: 127 SINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI--EYRFENKVI 184
S+N DR+LS R I + K +QLL V C+ IA+K EE P + E+ + + + +
Sbjct: 7 SVNLIDRYLSTRLIQKQK---LQLLGVTCMLIASKYEEMCAPRVEEFCFITDNTYTKEEV 177
Query: 185 KNMELLILSTLEWKMGLPTPFAYLHYF 211
ME +L + +++ +PT +L F
Sbjct: 178 LKMEREVLDLVHFQLSVPTIKTFLRRF 258
>BE348045 similar to PIR|T07672|T07 cyclin a2-type mitosis-specific -
soybean, partial (34%)
Length = 491
Score = 48.9 bits (115), Expect = 2e-06
Identities = 28/91 (30%), Positives = 49/91 (53%), Gaps = 2/91 (2%)
Frame = -1
Query: 123 TAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI--EYRFE 180
T YL++N DR LS+ + + + +QLL V C+ IA+K EE P + E+ + +
Sbjct: 485 TLYLTVNLIDRSLSQSLVQKQR---LQLLGVTCMLIASKYEEICAPRVEEFCFITDNTYT 315
Query: 181 NKVIKNMELLILSTLEWKMGLPTPFAYLHYF 211
+ ME +L+ L +++ +PT +L F
Sbjct: 314 KAEVLKMESEVLNLLHFQLSVPTTKTFLRRF 222
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,991,703
Number of Sequences: 63676
Number of extensions: 175249
Number of successful extensions: 1113
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 1081
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1094
length of query: 241
length of database: 12,639,632
effective HSP length: 94
effective length of query: 147
effective length of database: 6,654,088
effective search space: 978150936
effective search space used: 978150936
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146856.7


