

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146856.6 - phase: 0 /pseudo
(335 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
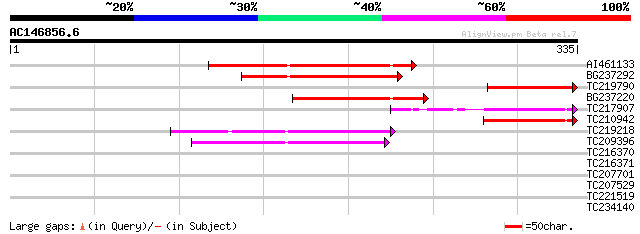
Score E
Sequences producing significant alignments: (bits) Value
AI461133 105 2e-23
BG237292 87 8e-18
TC219790 similar to GB|AAR24650.1|38603810|BT010872 At5g63160 {A... 74 1e-13
BG237220 67 1e-11
TC217907 similar to UP|Q9C5X9 (Q9C5X9) P300/CBP acetyltransferas... 65 3e-11
TC210942 62 3e-10
TC219218 weakly similar to UP|Q7F241 (Q7F241) Speckle-type POZ p... 55 4e-08
TC209396 weakly similar to GB|AAS77485.1|45825155|BT012360 At3g5... 46 2e-05
TC216370 32 0.31
TC216371 32 0.31
TC207701 similar to UP|Q84TR6 (Q84TR6) Subtilase, partial (6%) 28 5.8
TC207529 similar to UP|P93045 (P93045) SNF5 homolog BSH, partial... 28 5.8
TC221519 similar to UP|Q40372 (Q40372) Peroxidase precursor, par... 28 7.6
TC234140 27 10.0
>AI461133
Length = 384
Score = 105 bits (263), Expect = 2e-23
Identities = 57/123 (46%), Positives = 81/123 (65%)
Frame = +1
Query: 118 AVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVF 177
AV F+R+LYSS +EE+ ++ +HLL LSHVY V LK+ C + L LT +++VDV
Sbjct: 1 AVTAFVRFLYSSRCTEEEIDQYGMHLLALSHVYMVQQLKQRCIKGLT-HRLTTESVVDVL 177
Query: 178 QLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILESMIDEENSKKER 237
QLA LCDAP L L C + + KNF+ V +EGWK + + P LE +IL IDE ++K++
Sbjct: 178 QLARLCDAPDLHLRCMKLLAKNFKAVEATEGWKFLVKHDPWLELDILR-FIDEHETRKKK 354
Query: 238 IRK 240
R+
Sbjct: 355 SRR 363
>BG237292
Length = 425
Score = 87.4 bits (215), Expect = 8e-18
Identities = 45/95 (47%), Positives = 61/95 (63%)
Frame = +1
Query: 138 EFVLHLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKIL 197
++ +HLL LSHVY VP LK+ C + L LT +N+VDV QLA LCDAP L L C + +
Sbjct: 4 KYGMHLLALSHVYIVPQLKQRCIKGLT-HRLTTENVVDVLQLARLCDAPDLHLRCMKLLA 180
Query: 198 KNFRTVSESEGWKAMKQSHPVLEKEILESMIDEEN 232
NF+ V +EGWK + + P LE +IL + + EN
Sbjct: 181 NNFKAVEATEGWKFLXKHDPCLELDILRFIDEHEN 285
>TC219790 similar to GB|AAR24650.1|38603810|BT010872 At5g63160 {Arabidopsis
thaliana;} , partial (11%)
Length = 598
Score = 73.6 bits (179), Expect = 1e-13
Identities = 30/54 (55%), Positives = 40/54 (73%), Gaps = 1/54 (1%)
Frame = +3
Query: 283 YTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPD-FCRVPLC 335
+ +C+GL++L+RHFA C+ +V GGC CKRMWQL LHS +C D D C+VP C
Sbjct: 3 FATCQGLQVLIRHFATCEKKVRGGCVRCKRMWQLFRLHSYVCHDTDSSCKVPFC 164
>BG237220
Length = 450
Score = 67.0 bits (162), Expect = 1e-11
Identities = 32/80 (40%), Positives = 50/80 (62%)
Frame = +1
Query: 168 LTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILESM 227
L+ +N+VDV QL+ LCDAP L L C + + F+ V E+EGWK ++ P LE ++L +
Sbjct: 13 LSTENVVDVLQLSRLCDAPDLYLKCVKLLRNRFKAVKETEGWKFLESHDPWLELDVLR-L 189
Query: 228 IDEENSKKERIRKMNEREVY 247
+ E +K R+RK E + +
Sbjct: 190 MGELEKRKRRVRKWREEDTF 249
>TC217907 similar to UP|Q9C5X9 (Q9C5X9) P300/CBP acetyltransferase-related
protein 2, partial (14%)
Length = 1911
Score = 65.5 bits (158), Expect = 3e-11
Identities = 39/110 (35%), Positives = 58/110 (52%)
Frame = +1
Query: 226 SMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTS 285
SM D + KE R++ +QL ++ LVH + CR+ C+Y +
Sbjct: 421 SMADRDAQNKEA-RQLR----VIQLRKMLDLLVHASQ--CRSAH-----------CQYPN 546
Query: 286 CKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
C+ ++ L RH CK+R GGC CK+MW LL+LH+R C + + C VP C
Sbjct: 547 CRKVKGLFRHGMHCKIRASGGCVLCKKMWYLLQLHARACKESE-CHVPRC 693
>TC210942
Length = 823
Score = 62.4 bits (150), Expect = 3e-10
Identities = 26/55 (47%), Positives = 36/55 (65%)
Frame = +2
Query: 281 CRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
C+Y +C+ ++ L RH CK R GGC CK+MW LL+LH+R C + + C VP C
Sbjct: 71 CQYPNCRKVKGLFRHGMHCKTRASGGCVLCKKMWYLLQLHARACKESE-CHVPRC 232
>TC219218 weakly similar to UP|Q7F241 (Q7F241) Speckle-type POZ protein-like
protein, partial (59%)
Length = 837
Score = 55.1 bits (131), Expect = 4e-08
Identities = 38/133 (28%), Positives = 64/133 (47%)
Frame = +1
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
MLK R +I I V +D + F+ YLY++ E E +LLVL Y V HL
Sbjct: 244 MLKSDMTERRSGTIKISDVSYDTLHAFVNYLYTA--EASLDNELACNLLVLGEKYQVKHL 417
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
K CE+ L + + D + + A + +L + IL + +++++E + + +
Sbjct: 418 KTYCEKYL-IAKMNWDKAISNYAFAYQYNCKQLQSVSLAVILDHMDSLTQNECYAELVDT 594
Query: 216 HPVLEKEILESMI 228
+P L EI E+ I
Sbjct: 595 NPRLVVEIYETYI 633
>TC209396 weakly similar to GB|AAS77485.1|45825155|BT012360 At3g56230
{Arabidopsis thaliana;} , partial (44%)
Length = 1116
Score = 46.2 bits (108), Expect = 2e-05
Identities = 28/117 (23%), Positives = 56/117 (46%)
Frame = +2
Query: 108 SISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELGL 167
+I+I + H+ + + +LYS E++++ V L + Y +PHL + CE+ L L
Sbjct: 653 AITIPDLNHEELESLLEFLYSGTLNVEKLEKHVYALSQAADKYVIPHLLKHCERYL-LSS 829
Query: 168 LTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEIL 224
L+ N ++ ++A C L ++KN + S ++A P L +++
Sbjct: 830 LSTSNALETLEIADTCSNHNLKETTLNFLVKNIEHMVSSPKFEAFVHRSPHLTVQLV 1000
>TC216370
Length = 1349
Score = 32.3 bits (72), Expect = 0.31
Identities = 23/86 (26%), Positives = 43/86 (49%), Gaps = 2/86 (2%)
Frame = +3
Query: 142 HLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKIL--KN 199
HLL + Y + L+ CE L + ++ + LA +L +C + I +N
Sbjct: 645 HLLAAADRYGLERLRLMCEASL-CEDVAINTVATTLALAEQHHCFQLKAVCLKFIATSEN 821
Query: 200 FRTVSESEGWKAMKQSHPVLEKEILE 225
R V +++G++ +K+S P + E+LE
Sbjct: 822 LRAVMQTDGFEYLKESCPSVLTELLE 899
>TC216371
Length = 1127
Score = 32.3 bits (72), Expect = 0.31
Identities = 22/86 (25%), Positives = 43/86 (49%), Gaps = 2/86 (2%)
Frame = +1
Query: 142 HLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILK--N 199
HLL + Y + L+ CE L + ++ + LA +L +C + + + N
Sbjct: 526 HLLAAADRYGLERLRLMCETSL-CEDVAINTVATTLALAEQHHCFQLKAVCLKFVARPEN 702
Query: 200 FRTVSESEGWKAMKQSHPVLEKEILE 225
R V +++G++ +K+S P + E+LE
Sbjct: 703 LRAVMQTDGFEYLKESCPSVLTELLE 780
>TC207701 similar to UP|Q84TR6 (Q84TR6) Subtilase, partial (6%)
Length = 763
Score = 28.1 bits (61), Expect = 5.8
Identities = 12/41 (29%), Positives = 18/41 (43%)
Frame = +1
Query: 282 RYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSR 322
R++ K LEL H A L+ C H + W +H +
Sbjct: 133 RFSQRK*LELPFNHIAKSSLKTRNHCSHSHKCWATKHIHCK 255
>TC207529 similar to UP|P93045 (P93045) SNF5 homolog BSH, partial (82%)
Length = 1265
Score = 28.1 bits (61), Expect = 5.8
Identities = 9/18 (50%), Positives = 13/18 (72%)
Frame = -2
Query: 310 CKRMWQLLELHSRLCADP 327
C+R WQ L+ HS+ C+ P
Sbjct: 484 CRRFWQTLQGHSQSCSSP 431
>TC221519 similar to UP|Q40372 (Q40372) Peroxidase precursor, partial (70%)
Length = 746
Score = 27.7 bits (60), Expect = 7.6
Identities = 17/56 (30%), Positives = 23/56 (40%), Gaps = 12/56 (21%)
Frame = +2
Query: 281 CRYTSCKGLELLVRHFAGCKLRVPG------------GCGHCKRMWQLLELHSRLC 324
CRY SC L + GC + VP GCG C+ +L+L + C
Sbjct: 407 CRYLSCCS-S*LCSYIGGCTILVPSVTRQKRCNICKQGCGKCQSPSPILQLPTASC 571
>TC234140
Length = 526
Score = 27.3 bits (59), Expect = 10.0
Identities = 16/51 (31%), Positives = 23/51 (44%)
Frame = -3
Query: 181 LLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILESMIDEE 231
LLC+ P I HRKI K GWK + ++ + I+ +I E
Sbjct: 266 LLCEQPNYLTILHRKIFK----PHPKSGWKIPQPNYLTILHRIIFQLIMAE 126
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.342 0.149 0.512
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,614,110
Number of Sequences: 63676
Number of extensions: 310260
Number of successful extensions: 2237
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 2218
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2233
length of query: 335
length of database: 12,639,632
effective HSP length: 98
effective length of query: 237
effective length of database: 6,399,384
effective search space: 1516654008
effective search space used: 1516654008
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146856.6


