

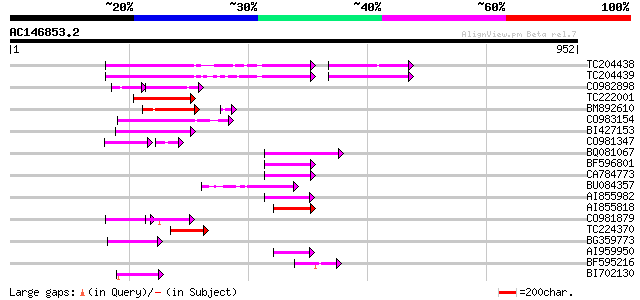
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146853.2 - phase: 0 /pseudo
(952 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 136 2e-34
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 136 1e-33
CO982898 77 4e-22
TC222001 similar to UP|C716_NEPRA (O04164) Cytochrome P450 71A6 ... 88 2e-17
BM892610 86 6e-17
CO983154 84 4e-16
BI427153 83 7e-16
CO981347 54 4e-14
BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa ... 75 1e-13
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 75 1e-13
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 70 6e-12
BU084357 69 1e-11
AI855982 66 8e-11
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 64 3e-10
CO981879 62 2e-09
TC224370 59 1e-08
BG359773 52 9e-07
AI959950 51 2e-06
BF595216 51 3e-06
BI702130 weakly similar to GP|4234854|gb| pol polyprotein {Zea m... 49 8e-06
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 136 bits (342), Expect(2) = 2e-34
Identities = 101/358 (28%), Positives = 157/358 (43%), Gaps = 5/358 (1%)
Frame = +1
Query: 161 NSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEM 220
NS F + C + GI S T QNG ERK R+ R +L LP + W A+
Sbjct: 2485 NSKFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNT 2664
Query: 221 ATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVF 280
A Y+ N + + + ++ + P+ H +FG C+ L + K+ P+S +F
Sbjct: 2665 ACYIHNRVTLRRGTPTTLYEIWKGRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIF 2844
Query: 281 LGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQH 340
LGY +N R Y+ + ++ +V+ ++ + P K + +
Sbjct: 2845 LGYSTNSRAYRVFNSRTRTVMESINVVVDD-LTPARKKDVEE------------------ 2967
Query: 341 LMDQTQTGPPIPQPAHQPNHI--TSPCPNSPNITSPPQSNPTSPIQQNLPP---IFQPTL 395
D +G + A + + + PNI P + P+ IQ+ P I P
Sbjct: 2968 --DVRTSGDNVADTAKSAENAENSDSATDEPNINQPDK-RPSIRIQKMHPKELIIGDP-- 3132
Query: 396 QANTKPITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKN 455
N TRS+ N + V+K P+N AL D W AM +E +N
Sbjct: 3133 --NRGVTTRSREIEIVSNSCF------VSKIE-PKNVKEALTDEFWINAMQEELEQFKRN 3285
Query: 456 KTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
+ W+LVPRP NVI + WIF++K +G R+KARLV G Q G+D ETF+P+
Sbjct: 3286 EVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPV 3459
Score = 29.3 bits (64), Expect(2) = 2e-34
Identities = 44/145 (30%), Positives = 64/145 (43%), Gaps = 3/145 (2%)
Frame = +2
Query: 536 WMSRMLFFMVN*KKQCTCTNLWDLKILKTPIMYAVFANHSMGLNKLLELGISDLSTTCHL 595
WM R F M * K+ ++ DL I IMY SM +KL ELG+ ++ L
Sbjct: 3527 WM*RARF*MDT*MKKPMWSSQRDL*IQLIQIMYTGSRRLSMD*SKLQELGMKG*QSSL-L 3703
Query: 596 LVFLKANVITLFSYTR---KTFI*HIIYSLLMILFSPRPQMICDNLSSHYSVLNLL*KIW 652
+ +T S + KT *H +LM L +M C ++ S+ LNL * +
Sbjct: 3704 SKGIGREELTRLSLSNKMLKT***H--RYMLMTLCLEGCRMRCFDILSNRCNLNLR*VLL 3877
Query: 653 DP*VTSWELQLLVTNKGCFSHKRSM 677
+ *+ W+ + SHK SM
Sbjct: 3878 ES*LIFWDSK*SRWKTPYSSHKASM 3952
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 136 bits (342), Expect(2) = 1e-33
Identities = 102/353 (28%), Positives = 159/353 (44%)
Frame = +1
Query: 161 NSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEM 220
NS F + C + GI S T QNG ERK R+ R +L LP + W A+
Sbjct: 2482 NSRFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNT 2661
Query: 221 ATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVF 280
A Y+ N + + + ++ + PS H +FG C+ L + K+ P+S +F
Sbjct: 2662 ACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIF 2841
Query: 281 LGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQH 340
LGY +N R Y+ + ++ +V+ ++ + P K + + + T DN
Sbjct: 2842 LGYSTNSRAYRVFNSRTRTVMESINVVVDD-LSPARKKDVEE-DVRTSGDN--------- 2988
Query: 341 LMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANTK 400
+ D ++G + A + T + NI P + + T Q + P N
Sbjct: 2989 VADAAKSG----ENAENSDSAT----DESNINQPDKRSSTRI--QKMHPKELIIGDPNRG 3138
Query: 401 PITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDL 460
TRS+ N + V+K P+N AL D W AM +E +N+ W+L
Sbjct: 3139 VTTRSREVEIVSNSCF------VSKIE-PKNVKEALTDEFWINAMQEELEQFKRNEVWEL 3297
Query: 461 VPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
VPRP NVI + WIF++K +G R+KARLV G Q G+D ETF+P+
Sbjct: 3298 VPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPV 3456
Score = 26.2 bits (56), Expect(2) = 1e-33
Identities = 39/143 (27%), Positives = 61/143 (42%), Gaps = 1/143 (0%)
Frame = +2
Query: 536 WMSRMLFFMVN*KKQCTCTNLWDLKILKTPIMYAVFANHSMGLNKLLELGISDLSTTCHL 595
WM R F M * K+ ++ DL+ IMY SM +KL ELG+ ++ L
Sbjct: 3524 WM*RAHF*MDT*MKKSMWSSQRDLQTRLIQIMYTGSRRLSMD*SKLQELGMKG*QSSL-L 3700
Query: 596 LVFLKANVITLFSYTRKTF-I*HIIYSLLMILFSPRPQMICDNLSSHYSVLNLL*KIWDP 654
+ +T S + K * + +LM L +M C ++ + LNL * + +
Sbjct: 3701 SKGIGREELTRPSLSNKMLKT**LHRYMLMTLCLEGCRMRCFDILFNRCNLNLR*VLLES 3880
Query: 655 *VTSWELQLLVTNKGCFSHKRSM 677
*+ W+ + SHK M
Sbjct: 3881 *LIFWDFK*SRWRTPYSSHKAGM 3949
>CO982898
Length = 816
Score = 76.6 bits (187), Expect(2) = 4e-22
Identities = 38/97 (39%), Positives = 56/97 (57%)
Frame = -1
Query: 229 PHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHR 288
P +INF P L+++ Y+ RVFGC CFPL I K +S +C+FLG ++H+
Sbjct: 597 PTVSINFAVPYTQLFNQMLDYSFPRVFGCACFPLIRPYNIQKFNFKSHECIFLGNYTSHK 418
Query: 289 GYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNT 325
YKCL ++ + V+FNE FPY +L +P +T
Sbjct: 417 NYKCLAPAGK--LVSKDVIFNEI*FPYKELFLPDSST 313
Score = 47.4 bits (111), Expect(2) = 4e-22
Identities = 27/59 (45%), Positives = 33/59 (55%)
Frame = -3
Query: 172 GIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPH 230
GI+ +L P+T QN ERK R +LL HASLP FW HA +L+N LPH
Sbjct: 766 GIIHKLIFPHTQYQN-VVERKQRHIVKFGLSLLSHASLPLKFWGHAFITPVFLINRLPH 593
>TC222001 similar to UP|C716_NEPRA (O04164) Cytochrome P450 71A6 (Fragment)
, partial (21%)
Length = 912
Score = 87.8 bits (216), Expect = 2e-17
Identities = 41/103 (39%), Positives = 63/103 (60%)
Frame = -2
Query: 209 LPSSFWHHALEMATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKI 268
+P +FW++AL A YL+N +P + SP + L+ P +HLR+FGCLC+ +
Sbjct: 911 MPPNFWNYALLHAAYLINCIPTPFLQNTSPYERLHGHIPDISHLRIFGCLCYASTIKANR 732
Query: 269 YKLQPRSTKCVFLGYPSNHRGYKCLGLLSNKIIICRHVLFNEN 311
KL+PR+ C+F+G+ N +GY L S+ II R+V+F EN
Sbjct: 731 KKLEPRAHPCIFIGFKPNTKGYMLYDLHSHNIITSRNVVFYEN 603
>BM892610
Length = 421
Score = 85.5 bits (210), Expect(2) = 6e-17
Identities = 42/96 (43%), Positives = 62/96 (63%)
Frame = -3
Query: 223 YLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLG 282
YL+N LP ++ F+ P V ++K P+YN LR FGC C+P +KL RS +C+FLG
Sbjct: 413 YLINRLPTTSLKFQVPCTV-FNKLPNYNFLRNFGCSCYPFLRPYNKHKLDFRSHECLFLG 237
Query: 283 YPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKL 318
Y + H+GYKCL + K+ + + V+FNE +PY+ L
Sbjct: 236 YSTPHKGYKCLS-PNGKLYVSKDVIFNELKYPYSDL 132
Score = 21.2 bits (43), Expect(2) = 6e-17
Identities = 11/27 (40%), Positives = 14/27 (51%)
Frame = -2
Query: 355 AHQPNHITSPCPNSPNITSPPQSNPTS 381
AH P + SP PN P + +N TS
Sbjct: 78 AHIP--LVSPSPNKPTPSQHAGTNSTS 4
>CO983154
Length = 568
Score = 83.6 bits (205), Expect = 4e-16
Identities = 55/196 (28%), Positives = 91/196 (46%), Gaps = 1/196 (0%)
Frame = +3
Query: 181 YTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFESPLK 240
+T QNG +ERK R R+L+++ ++P W A+ + +L+N +P ++ + P
Sbjct: 3 HTPQQNGIAERKNRHLLETARSLMLNLNVPIHHWGDAVLTSCFLINRMPSSSLENQIPHS 182
Query: 241 VLYHKDPSYN-HLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGYKCLGLLSNK 299
+++ DP ++ +VFGC CF S + KL RS KCVFLGY +GYKC +
Sbjct: 183 LVFPHDPLFHVSPKVFGCTCFVHDLSPGLDKLSARSVKCVFLGYSRLQKGYKCYSPTMRR 362
Query: 300 IIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQHLMDQTQTGPPIPQPAHQPN 359
+ V F E+ P+ + ++ + SPY P+ +
Sbjct: 363 YYMSADVTFFEDT-PFFSPSVDHSSSLQEVLPIPSPY-------------PLXNSGQNVS 500
Query: 360 HITSPCPNSPNITSPP 375
+ S PNS + PP
Sbjct: 501 IVPSSSPNSLEVILPP 548
>BI427153
Length = 422
Score = 82.8 bits (203), Expect = 7e-16
Identities = 45/135 (33%), Positives = 73/135 (53%), Gaps = 1/135 (0%)
Frame = +1
Query: 178 SCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFES 237
+CP+T QNG +ERK R+L++++++P+ W A+ A +L+N +P ++ +
Sbjct: 10 TCPHTPQQNGIAERKNHHLLETARSLMLNSNVPTHHWGDAVLTACFLINRMPSSSLENQI 189
Query: 238 PLKVLYHKD-PSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGYKCLGLL 296
P +++ D Y +VFGC CF S + KL RS KCVFLGY +GY C
Sbjct: 190 PHSIVFPNDLLFYVSPKVFGCTCFVHDLSPGLDKLSARSVKCVFLGYSRLQKGYTCYFPN 369
Query: 297 SNKIIICRHVLFNEN 311
+ + +V F E+
Sbjct: 370 MRRYYMSANVTFFED 414
>CO981347
Length = 624
Score = 54.3 bits (129), Expect(2) = 4e-14
Identities = 27/81 (33%), Positives = 42/81 (51%)
Frame = +2
Query: 160 VNSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALE 219
V F + C GI P+T QNG +ER + +R +L+ A LP +FW A
Sbjct: 197 VLEQFNEFCRKIGIKRHKIVPHTP*QNGLAERMNMTILERVRCMLLSARLPKTFWGEAAN 376
Query: 220 MATYLMNILPHKTINFESPLK 240
+YL+N P T+ F++P++
Sbjct: 377 TTSYLINRCPSSTLGFKTPME 439
Score = 42.7 bits (99), Expect(2) = 4e-14
Identities = 22/47 (46%), Positives = 28/47 (58%)
Frame = +3
Query: 245 KDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGYK 291
K P+Y+ L+VFG L F K KL R+ KCVF+GYP + YK
Sbjct: 453 KPPNYSGLKVFGSLAFD---HVKQGKLDARAVKCVFIGYPKGVKRYK 584
>BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa (japonica
cultivar-group)}, partial (18%)
Length = 430
Score = 75.1 bits (183), Expect = 1e-13
Identities = 47/132 (35%), Positives = 66/132 (49%)
Frame = +1
Query: 429 PRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFER 488
P AL P W+ AM ++++L++NKT L P I W+FR KE GT R
Sbjct: 34 PSTVKQALISPPWRQAMQADFDALMENKTLTLTSLPSGKAAIDCKWVFRIKENLYGTLNR 213
Query: 489 HKARLVGDGAGQQVGIDCGETFSPM*NQLLFAQFSELLHPDLGKFISWMSRMLFFMVN*K 548
+++RLV G + G D ETFSP+ LF F +G F + MLF+M *
Sbjct: 214 YRSRLVAKGFHLKFGCDYSETFSPVIEL*LFDSFCS*PSLIIGHFNK*ILIMLFYMDF*L 393
Query: 549 KQCTCTNLWDLK 560
++CT + LK
Sbjct: 394 RRCTWFSYQGLK 429
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 75.1 bits (183), Expect = 1e-13
Identities = 35/85 (41%), Positives = 50/85 (58%)
Frame = +3
Query: 429 PRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFER 488
P+N A+ D NW + M +E N +N W LV +P + VI + W+FR+K G R
Sbjct: 6 PKNIKEAIVDDNWIIVMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIR 185
Query: 489 HKARLVGDGAGQQVGIDCGETFSPM 513
+KARLV G Q+ GID ET++P+
Sbjct: 186 NKARLVAKGYNQEEGIDYEETYAPV 260
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 69.7 bits (169), Expect = 6e-12
Identities = 35/86 (40%), Positives = 47/86 (53%)
Frame = +3
Query: 428 LPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFE 487
+P AL P W+ AM DE +L N TW+LVP PP + W++ K +G +
Sbjct: 18 VPSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGPNGKVD 197
Query: 488 RHKARLVGDGAGQQVGIDCGETFSPM 513
R KARLV G Q GI+ +TFSP+
Sbjct: 198 RLKARLVAKGYTQVYGIEYCDTFSPV 275
>BU084357
Length = 431
Score = 68.9 bits (167), Expect = 1e-11
Identities = 55/165 (33%), Positives = 79/165 (47%), Gaps = 1/165 (0%)
Frame = +1
Query: 322 QPNTYTFLDNELSPYIIQHLMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPTS 381
Q T + L++ LSP ++QTQT PN + N+ S +N +
Sbjct: 1 QHQTTSNLNSTLSP------LNQTQT----------PNEESQSNSNNLYTCSSLPANANA 132
Query: 382 PIQQNLPPIFQPTLQANTKP-ITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPN 440
LPP + +L +T P +TRS++GIFKP Q + P A+K
Sbjct: 133 -----LPPCSKTSL--STYPMVTRSKNGIFKPKQVHLATKFPFPSLVEPTCVTQAMKHLE 291
Query: 441 WKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGT 485
WK M +E N+LI N TW LVP ++NVI + W FR K +DG+
Sbjct: 292 WKKTMLEELNALIANGTWTLVPS*LNLNVIGNKWDFRLKHNTDGS 426
>AI855982
Length = 484
Score = 65.9 bits (159), Expect = 8e-11
Identities = 32/83 (38%), Positives = 46/83 (54%)
Frame = +2
Query: 429 PRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFER 488
P+N A+ D NW +AM +E N +N W LV +P + VI + W+FR+K
Sbjct: 101 PKNIKEAIVDDNWIIAMQEELNQFERNNVWKLVEKPDNYPVI*TKWVFRNKLDEHRIIII 280
Query: 489 HKARLVGDGAGQQVGIDCGETFS 511
HKARLV +G Q G+D T++
Sbjct: 281 HKARLVAEGYNQVDGLDYEHTYA 349
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 63.9 bits (154), Expect = 3e-10
Identities = 30/71 (42%), Positives = 43/71 (60%)
Frame = -3
Query: 443 MAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQV 502
+AM +E N +N W LV +P + VI + W+FR+K G R+KARLV G Q+
Sbjct: 461 IAMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEE 282
Query: 503 GIDCGETFSPM 513
GID ET++P+
Sbjct: 281 GIDYEETYAPV 249
>CO981879
Length = 576
Score = 61.6 bits (148), Expect = 2e-09
Identities = 33/88 (37%), Positives = 44/88 (49%), Gaps = 5/88 (5%)
Frame = -2
Query: 228 LPHKTINFESPLKVLYHKDPSYN-----HLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLG 282
+P K +NF +PL V P+ L++FGC F KL+PR+ KCVF+G
Sbjct: 281 MPSKILNFRTPLDVFTSAFPNNRLSCTLPLKIFGCTVFVHIHEPNQGKLEPRAKKCVFVG 102
Query: 283 YPSNHRGYKCLGLLSNKIIICRHVLFNE 310
Y N +GYKC S K + V F E
Sbjct: 101 YAPNQKGYKCFDPTSKKTFVTIDVTFFE 18
Score = 50.4 bits (119), Expect = 4e-06
Identities = 28/85 (32%), Positives = 41/85 (47%)
Frame = -1
Query: 161 NSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEM 220
N + K NGI+ + SC T QNG +ERK R + R LL P W A+
Sbjct: 483 NKHLSKXXLENGIIHQSSCVDTPQQNGVAERKNRHLXEVARALLFQNKAPKYXWGEAILT 304
Query: 221 ATYLMNILPHKTINFESPLKVLYHK 245
TYL N + + F++ ++ +HK
Sbjct: 303 GTYLKNKNA*QNLEFQNSIR-CFHK 232
>TC224370
Length = 320
Score = 58.5 bits (140), Expect = 1e-08
Identities = 31/65 (47%), Positives = 41/65 (62%)
Frame = -2
Query: 270 KLQPRSTKCVFLGYPSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFL 329
KL RS +C+FLGY S+H+GYKCL + +I I + VLFNE FPY+ L N+ L
Sbjct: 295 KLNFRSQECLFLGYSSSHKGYKCLSSI-GRIYISKDVLFNEIRFPYSDLFASSSNSI*NL 119
Query: 330 DNELS 334
D+ S
Sbjct: 118 DSYFS 104
>BG359773
Length = 382
Score = 52.4 bits (124), Expect = 9e-07
Identities = 28/93 (30%), Positives = 54/93 (57%)
Frame = -1
Query: 164 FQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATY 223
F K+ + GI + + NG SE++ R +++ ++LI+ +LP S W +AL++ Y
Sbjct: 322 FAKLIQKRGICAQYTMLGIPQ*NGVSEKRNRILMDMVTSMLINLTLPISLWMYALKIVMY 143
Query: 224 LMNILPHKTINFESPLKVLYHKDPSYNHLRVFG 256
L+N +P K + ++ ++ ++ PS HL V+G
Sbjct: 142 LLNRVPSKAVP-KTHFELWTNRTPSIRHLDVWG 47
>AI959950
Length = 466
Score = 51.2 bits (121), Expect = 2e-06
Identities = 29/68 (42%), Positives = 36/68 (52%)
Frame = -1
Query: 444 AMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVG 503
AM +E + KN LV P V+ WIF +K DG R+KARLV G QQ G
Sbjct: 391 AMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQEG 212
Query: 504 IDCGETFS 511
ID +TF+
Sbjct: 211 IDYPKTFA 188
>BF595216
Length = 421
Score = 50.8 bits (120), Expect = 3e-06
Identities = 35/82 (42%), Positives = 45/82 (54%), Gaps = 4/82 (4%)
Frame = +1
Query: 479 KEKSDGTFERHKARLVGDGAGQQVGIDCGETFS----PM*NQLLFAQFSELLHPDLGKFI 534
KE SDGT ++KA+LV G QQ G D ETFS P+ +LL + P G F
Sbjct: 124 KENSDGTVNKYKAKLVAKGFHQQYGTDYIETFSLVIKPITMRLLLT--LAVT*PITGPFS 297
Query: 535 SWMSRMLFFMVN*KKQCTCTNL 556
SWM M F MV K++ C++L
Sbjct: 298 SWM*IMPFLMVFYKRKSICSSL 363
>BI702130 weakly similar to GP|4234854|gb| pol polyprotein {Zea mays},
partial (14%)
Length = 421
Score = 49.3 bits (116), Expect = 8e-06
Identities = 25/83 (30%), Positives = 47/83 (56%), Gaps = 4/83 (4%)
Frame = -1
Query: 179 CP----YTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTIN 234
CP + S +ER+ R+ +++R++ + LP W AL+ A Y++N +P K ++
Sbjct: 289 CPIHYAWFSGSEWVAERRNRTLLDMVRSMRSNVKLPQFLWIDALKTAAYILNRVPTKAVS 110
Query: 235 FESPLKVLYHKDPSYNHLRVFGC 257
++P ++ PS H+RV+GC
Sbjct: 109 -KTPFELFKGWKPSLRHIRVWGC 44
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.337 0.147 0.487
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,251,132
Number of Sequences: 63676
Number of extensions: 961715
Number of successful extensions: 12093
Number of sequences better than 10.0: 286
Number of HSP's better than 10.0 without gapping: 10067
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11404
length of query: 952
length of database: 12,639,632
effective HSP length: 106
effective length of query: 846
effective length of database: 5,889,976
effective search space: 4982919696
effective search space used: 4982919696
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146853.2


