

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146842.6 - phase: 0
(535 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
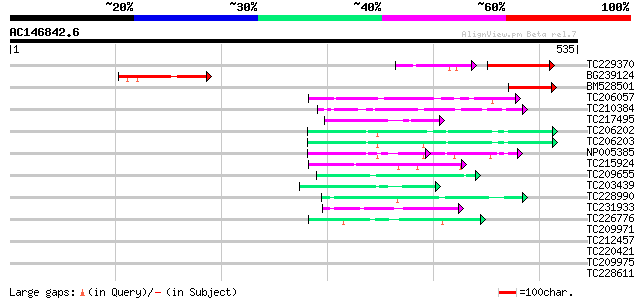
Score E
Sequences producing significant alignments: (bits) Value
TC229370 similar to UP|Q9LSA0 (Q9LSA0) Emb|CAB62463.1, partial (7%) 61 4e-20
BG239124 92 6e-19
BM528501 61 1e-09
TC206057 homologue to UP|Q9ZRX1 (Q9ZRX1) Cytosolic chaperonin, d... 47 2e-05
TC210384 weakly similar to UP|Q8VZF9 (Q8VZF9) AT5g12890/T24H18_6... 46 4e-05
TC217495 similar to UP|Q9ASU0 (Q9ASU0) At1g26110/F28B23_21, part... 45 1e-04
TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragme... 44 1e-04
TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragme... 44 2e-04
NP005385 DNA-directed RNA polymerase 44 2e-04
TC215924 similar to GB|AAS68109.1|45592910|BT011785 At2g38360 {A... 42 5e-04
TC209655 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partia... 42 5e-04
TC203439 weakly similar to UP|Q9LX02 (Q9LX02) ESTs AU082316(E336... 42 5e-04
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 42 7e-04
TC231933 similar to PIR|F86340|F86340 protein F2D10.34 [imported... 42 7e-04
TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {A... 42 7e-04
TC209971 similar to UP|Q9MA24 (Q9MA24) T5E21.9, partial (73%) 41 0.002
TC212457 UP|Q39871 (Q39871) Maturation polypeptide, complete 41 0.002
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 40 0.002
TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%) 40 0.002
TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxida... 40 0.002
>TC229370 similar to UP|Q9LSA0 (Q9LSA0) Emb|CAB62463.1, partial (7%)
Length = 1134
Score = 61.2 bits (147), Expect(2) = 4e-20
Identities = 32/64 (50%), Positives = 40/64 (62%), Gaps = 1/64 (1%)
Frame = +3
Query: 452 TPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSR-SYQDEFDDTDFTCPFDVDDDDTT 510
T +K KE G ++ + SP+I FSR+SSR S+QDE DD DF+CPFDVDD D
Sbjct: 387 TGQKFVRDSKEDSGRFSGLLSSSDSPRIGFSRTSSRLSFQDELDDGDFSCPFDVDDVDPP 566
Query: 511 DPGS 514
D S
Sbjct: 567 DAQS 578
Score = 55.1 bits (131), Expect(2) = 4e-20
Identities = 35/83 (42%), Positives = 47/83 (56%), Gaps = 7/83 (8%)
Frame = +2
Query: 365 NFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEV-----TYSPGH 419
++DEY SP FSPSPSP S P Y G KT + SE+APV IP++ + SP
Sbjct: 101 SYDEYQLSPPFSPSPSP--SPPTYFCGGNPMKTRMCSETAPVTIPHSVMGKSSRNLSPNF 274
Query: 420 T--SRQNLPPSTPIRISRCTSET 440
+ SR +LPP +P R + E+
Sbjct: 275 SDPSRNSLPPLSPRRTDGSSQES 343
>BG239124
Length = 317
Score = 92.0 bits (227), Expect = 6e-19
Identities = 56/97 (57%), Positives = 68/97 (69%), Gaps = 9/97 (9%)
Frame = +3
Query: 103 LSFSPKV----RSFVKERNPF-----EEFGGGSEQNEKIVERWVIQYESKKIKDCSSSNT 153
+SFSPK S +KER PF +E G ++EKIVERW++QYES+K +D +S +
Sbjct: 42 VSFSPKRVLPRSSSLKERCPFGWNTDQEELGVVGRSEKIVERWLVQYESRKTRDSNSGS- 218
Query: 154 ANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAFK 190
RRSSN L NLYKKSTLLLRSLYATVRLLPA+K
Sbjct: 219 ----RRSSNVSLHNLYKKSTLLLRSLYATVRLLPAYK 317
>BM528501
Length = 425
Score = 60.8 bits (146), Expect = 1e-09
Identities = 29/47 (61%), Positives = 36/47 (75%), Gaps = 1/47 (2%)
Frame = -1
Query: 471 IAPNSSPQISFSRSSSR-SYQDEFDDTDFTCPFDVDDDDTTDPGSRY 516
++ + SP+I FSR+SSR S+QDE DD DF+CPFDVDD D D SRY
Sbjct: 422 LSSSDSPRIGFSRTSSRLSFQDELDDGDFSCPFDVDDVDPPDAQSRY 282
>TC206057 homologue to UP|Q9ZRX1 (Q9ZRX1) Cytosolic chaperonin,
delta-subunit, complete
Length = 1930
Score = 47.4 bits (111), Expect = 2e-05
Identities = 57/202 (28%), Positives = 86/202 (42%), Gaps = 2/202 (0%)
Frame = +3
Query: 283 SLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQG 342
S P+ PS SS P R S S + R PPA S + + S + S++S
Sbjct: 267 SSPMTAPPSSTRCRSSSPPPRCSS-SSPSPRTPPPATAPLPSSSSLAPSSSSASSSSPTA 443
Query: 343 YCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSE 402
PPSS P+ + L + F +PSP+ SP+ +PS S P + + +S RS
Sbjct: 444 STPPSS----PTPSTRLP*RPSTFSPPWPSPSSSPTATPS*SPPAHPSTARSSANTPRS- 608
Query: 403 SAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPE--KMFSIG 460
++P+ +SP +PPS IS + + RN TTP K+ S+
Sbjct: 609 ----SLPSPSTPFSPSW-----MPPSPTWSIS---AMSRS*RNSAAPSTTPSS*KVSSLT 752
Query: 461 KESQKYSGGKIAPNSSPQISFS 482
+ S G A + SF+
Sbjct: 753 RRSATPPGDPPAWRTPRSPSFN 818
>TC210384 weakly similar to UP|Q8VZF9 (Q8VZF9) AT5g12890/T24H18_60, partial
(11%)
Length = 860
Score = 46.2 bits (108), Expect = 4e-05
Identities = 63/200 (31%), Positives = 89/200 (44%), Gaps = 2/200 (1%)
Frame = +3
Query: 291 SHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLP 350
SH + PS P S+ S S AS P +PSPT S S ASS+ PPS+ P
Sbjct: 300 SHQTPPS--PSSK--SPSLPPTTASLPTRRTPTPSPTTS------SFASSKPP-PPSNQP 446
Query: 351 PHPSEMSLLHK-KNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIP 409
PS + K KN+NF S + P PSP +S+ S +S + ++P P
Sbjct: 447 SKPSSKTSSSKTKNINF*SSPTSSSAGPPPSPRNSA-----SSTSSSAAPPASASPATTP 611
Query: 410 NAEVTYSPGHTSRQNLPPSTPIRIS-RCTSETERSRNLMQSCTTPEKMFSIGKESQKYSG 468
+ ++ + T P S+P RIS + S T RS + T P++M K
Sbjct: 612 SGTISPTAAST-----PTSSPYRISPKRVSFTARSYPI----TFPKQMAPTRGLCFK--- 755
Query: 469 GKIAPNSSPQISFSRSSSRS 488
I PN S + F + S+S
Sbjct: 756 KVIFPNGSTRTGFCSTPSKS 815
>TC217495 similar to UP|Q9ASU0 (Q9ASU0) At1g26110/F28B23_21, partial (59%)
Length = 2299
Score = 44.7 bits (104), Expect = 1e-04
Identities = 36/114 (31%), Positives = 50/114 (43%), Gaps = 1/114 (0%)
Frame = +2
Query: 298 SLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNAS-SQGYCPPSSLPPHPSEM 356
S+P S + Y N P V+ P S S S + PPS+LPP PS +
Sbjct: 710 SMPSSMQQPMQYPNITPPLPTVSSNLPELPSSLLPASASTPSITSASLPPSNLPPAPSAL 889
Query: 357 SLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPN 410
+ P+P+ P P+PS+ SP S S A L SE+ PV++ N
Sbjct: 890 A-------------PAPSALP-PAPSALSPSSSALSPAPSATLASENLPVSVTN 1009
>TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragment) ,
complete
Length = 1826
Score = 44.3 bits (103), Expect = 1e-04
Identities = 67/242 (27%), Positives = 97/242 (39%), Gaps = 6/242 (2%)
Frame = +3
Query: 282 PSLPVVRMPSHGSSPSSLPFS-RRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASS 340
P+ P S G SP+S +S +S + SP + +Y SP++S + S +S
Sbjct: 699 PTSPGYSPTSPGYSPTSPTYSPSSPGYSPTSPAYSPTSPSYSPTSPSYSPTSPSYS-PTS 875
Query: 341 QGYCP--PSSLPPHPSEMSLLHKKNVNFDEYYP-SPNFSP-SPSPSSSSPIYSLGSLASK 396
Y P PS P P+ + Y P SP++SP SPS S +SP YS S +
Sbjct: 876 PSYSPTSPSYSPTSPAYSPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPS-- 1049
Query: 397 TLLRSESAPVNIPNAEVTYSPGHTSRQNLPPS-TPIRISRCTSETERSRNLMQSCTTPEK 455
S ++P P + YSP S PS +P S + S +L S ++P
Sbjct: 1050---YSPTSPAYSPTSP-GYSPTSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRL 1217
Query: 456 MFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDDTDFTCPFDVDDDDTTDPGSR 515
S YS +PN SP +S SY + P++ P S
Sbjct: 1218-----SPSSPYS--PTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSGVSPDYSPSSP 1376
Query: 516 YF 517
F
Sbjct: 1377QF 1382
Score = 38.9 bits (89), Expect = 0.006
Identities = 48/159 (30%), Positives = 60/159 (37%), Gaps = 15/159 (9%)
Frame = +3
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
S PT+P Y SP + P+ P S G SP+S +S
Sbjct: 984 SYSPTSPSYSPTSPSY--SPTSPSYS--PTSPAYSPTSPGYSPTSPSYSPTSPSYSPTSP 1151
Query: 314 ASPPAVNYYSPSPTHSESHTLVSNAS-----SQGYCP--PSSLPPHPS--------EMSL 358
+ P YSPS +S S +S +S S Y P PS P PS S
Sbjct: 1152 SYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSS 1331
Query: 359 LHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKT 397
+ V+ D SP FSPS S S P YS S + T
Sbjct: 1332 PYNSGVSPDYSPSSPQFSPSTGYSPSQPGYSPSSTSQYT 1448
>TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragment) , partial
(96%)
Length = 2604
Score = 43.9 bits (102), Expect = 2e-04
Identities = 67/244 (27%), Positives = 98/244 (39%), Gaps = 8/244 (3%)
Frame = +1
Query: 282 PSLPVVRMPSHGSSPSSLPFS-RRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASS 340
P+ P S G SP+S +S +S + SP + +Y SP++S + S +S
Sbjct: 1156 PTSPGYSPTSPGYSPTSPTYSPSSPGYSPTSPAYSPTSPSYSPTSPSYSPTSPSYS-PTS 1332
Query: 341 QGYCP--PSSLPPHPSEMSLLHKKNVNFDEYYP-SPNFSP-SPSPSSSSPIYS--LGSLA 394
Y P PS P P+ + Y P SP++SP SPS S +SP YS S +
Sbjct: 1333 PSYSPTSPSYSPTSPAYSPTSPAYSPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYS 1512
Query: 395 SKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPS-TPIRISRCTSETERSRNLMQSCTTP 453
+ S ++P P + YSP S PS +P S + S +L S ++P
Sbjct: 1513 PTSPSYSPTSPAYSPTSP-GYSPTSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSP 1689
Query: 454 EKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDDTDFTCPFDVDDDDTTDPG 513
S YS +PN SP +S SY + P++ P
Sbjct: 1690 RL-----SPSSPYS--PTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSGVSPDYSPS 1848
Query: 514 SRYF 517
S F
Sbjct: 1849 SPQF 1860
Score = 38.9 bits (89), Expect = 0.006
Identities = 48/159 (30%), Positives = 60/159 (37%), Gaps = 15/159 (9%)
Frame = +1
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
S PT+P Y SP + P+ P S G SP+S +S
Sbjct: 1462 SYSPTSPSYSPTSPSY--SPTSPSYS--PTSPAYSPTSPGYSPTSPSYSPTSPSYSPTSP 1629
Query: 314 ASPPAVNYYSPSPTHSESHTLVSNAS-----SQGYCP--PSSLPPHPS--------EMSL 358
+ P YSPS +S S +S +S S Y P PS P PS S
Sbjct: 1630 SYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSS 1809
Query: 359 LHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKT 397
+ V+ D SP FSPS S S P YS S + T
Sbjct: 1810 PYNSGVSPDYSPSSPQFSPSTGYSPSQPGYSPSSTSQYT 1926
>NP005385 DNA-directed RNA polymerase
Length = 2934
Score = 43.5 bits (101), Expect = 2e-04
Identities = 39/117 (33%), Positives = 53/117 (44%), Gaps = 1/117 (0%)
Frame = +1
Query: 282 PSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQ 341
P+ P S +P S +S ++S + R SP + NY SP++S + S SS
Sbjct: 2596 PTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPTSPNYSPTSPSYSPTSPSYS-PSSP 2772
Query: 342 GYCPPSSLPPHPSEMSLLHKKNVNFDEYYP-SPNFSPSPSPSSSSPIYSLGSLASKT 397
Y P S P+ S +S +Y P SP FSPS S S P YS S + T
Sbjct: 2773 TYSPSS---PYNSGVS---------PDYSPTSPQFSPSAGYSPSQPGYSPSSTSQDT 2907
Score = 43.5 bits (101), Expect = 2e-04
Identities = 62/215 (28%), Positives = 92/215 (41%), Gaps = 12/215 (5%)
Frame = +1
Query: 282 PSLPVVRMPSHGSSPSSLPFSRRH-SWSYENCRASPPAVNYYSPSPTHSESHTLVSNASS 340
P+ P S G SP+S +S +S + SP + +Y SP +S + S+ +S
Sbjct: 2239 PTSPGYSPTSPGYSPTSPTYSPSSPGYSPTSPAYSPTSPSYSPTSPAYSPTSPAYSS-TS 2415
Query: 341 QGYCP--PSSLPPHPSEMSLLHKKNVNFDEYYP-SPNFSP-SPSPSSSSPIYS--LGSLA 394
Y P PS P P+ ++ Y P SP++SP SPS S +SP YS S +
Sbjct: 2416 PAYSPTSPSYSPTSPAYSPTSPSYSLTSPSYSPTSPSYSPTSPSYSPTSPAYSPTSPSYS 2595
Query: 395 SKTLLRSESAPVNIPNAEVTYSPG---HTSRQNLPPSTPIRISRCTSETERSRNLMQSCT 451
+ S ++P P + YSP S L P++P S + S + S
Sbjct: 2596 PTSPSYSPTSPSYNPQS-AKYSPSLAYSPSSPRLSPTSPNYSPTSPSYSPTSPSYSPSSP 2772
Query: 452 T--PEKMFSIGKESQKYSGGKIAPNSSPQISFSRS 484
T P ++ G S YS +P SP +S S
Sbjct: 2773 TYSPSSPYNSG-VSPDYS--PTSPQFSPSAGYSPS 2868
>TC215924 similar to GB|AAS68109.1|45592910|BT011785 At2g38360 {Arabidopsis
thaliana;} , partial (68%)
Length = 947
Score = 42.4 bits (98), Expect = 5e-04
Identities = 48/165 (29%), Positives = 67/165 (40%), Gaps = 16/165 (9%)
Frame = +3
Query: 283 SLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQG 342
S R S G++P S S + R SP + + +PS HS S +L S S
Sbjct: 195 STSAARGRSSGTAPRSRSPSPSPKPPSASARTSPTSASTTTPS-FHSFSQSLSSPILSLS 371
Query: 343 YCPPSSLPPHPSEMSLLHKKNVN------FDEYYPSPNFSPSPSPSS--------SSPIY 388
+S PP PS S + + N + F P + PSPS SS SSP+
Sbjct: 372 SSSSASSPPGPSSTSSVRQINHSLFLAELFQISKP*RSSPPSPSSSSSSPALDLFSSPLS 551
Query: 389 SLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQN--LPPSTPI 431
L S +S + RS + T P +S + LPP P+
Sbjct: 552 CLASPSSASTARSACRRICFLTTRTTLRPPDSSPSSAALPPPPPL 686
>TC209655 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partial (32%)
Length = 832
Score = 42.4 bits (98), Expect = 5e-04
Identities = 40/155 (25%), Positives = 62/155 (39%)
Frame = +1
Query: 290 PSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSL 349
P+ SS + S + + S P+ + P PT + + L +++ PSS
Sbjct: 7 PNQSSSKKAPTTSSPSTTAATTSPPSSPSSPTHRPLPTRTPTQPLPPHSAQ-----PSSN 171
Query: 350 PPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIP 409
P P+ S LH + P P SPS +PS ++ + + + S S P P
Sbjct: 172 SPSPTAPSRLHSGTRSSSPSAPPPPPSPSRTPSFHGSRNTISASPTSSSTPSSSTPSAAP 351
Query: 410 NAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSR 444
+ S + R +LP T R S ET SR
Sbjct: 352 KSSTKPSYSPSVRSSLPSPT-TRSSARAPETATSR 453
Score = 40.4 bits (93), Expect = 0.002
Identities = 42/166 (25%), Positives = 64/166 (38%)
Frame = +1
Query: 232 PVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPS 291
P + +S K + + +S P++P SP PL R P P PS
Sbjct: 4 PPNQSSSKKAPTTSSPSTTAATTSPPSSPSSP------THRPLPTRTPTQPLPPHSAQPS 165
Query: 292 HGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPP 351
+SPS SR HS + + ++PP S +P+ S +S + + P SS P
Sbjct: 166 -SNSPSPTAPSRLHSGTRSSSPSAPPPPPSPSRTPSFHGSRNTISASPTSSSTPSSSTPS 342
Query: 352 HPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKT 397
+ S P++SPS S SP + A +T
Sbjct: 343 AAPKSS-------------TKPSYSPSVRSSLPSPTTRSSARAPET 441
>TC203439 weakly similar to UP|Q9LX02 (Q9LX02) ESTs AU082316(E3368), partial
(27%)
Length = 1497
Score = 42.4 bits (98), Expect = 5e-04
Identities = 41/134 (30%), Positives = 54/134 (39%), Gaps = 1/134 (0%)
Frame = -2
Query: 274 LTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSY-ENCRASPPAVNYYSPSPTHSESH 332
L D L R + PS SPS PFS +H S E C S + P S S
Sbjct: 695 LYDHLARESPQCAIHFPSSSPSPSPSPFSSQHPHSAPETCTFSSSHSSPLFP*SQPSISR 516
Query: 333 TLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGS 392
+ +++ PP LP HP+ S YPS S S SSSS +
Sbjct: 515 SWQHPSATSSCFPP--LPQHPTSRS------------YPSAPSPASSSSSSSSLLPQRSP 378
Query: 393 LASKTLLRSESAPV 406
L + T R+ S+P+
Sbjct: 377 LPTCTSYRTHSSPL 336
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 42.0 bits (97), Expect = 7e-04
Identities = 52/203 (25%), Positives = 81/203 (39%), Gaps = 9/203 (4%)
Frame = +2
Query: 295 SPSSLPFSRRHSWSYENCRASP-PAVNYYSPSPTHSE-SHTLVSNASSQGYCPPSSLPPH 352
SP+ P + S ++ + R SP P ++ +PSP S S S+ S + P SLP
Sbjct: 320 SPALYPMT---SPTFRSSRTSPSPTISSPAPSPPLSPPSPPSASSTSPTTFSTPPSLPSS 490
Query: 353 PSEMSLLHKKNV------NF-DEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAP 405
+ +L + NF P P+ + S ++SSP SL S A + + +P
Sbjct: 491 IASPTLRFSTSTTTT*PENFLSPSLPCPSSATFTSAATSSPARSLPSTAPGSTSSTSPSP 670
Query: 406 VNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQK 465
A + +P S + PPS + T SR +++C T
Sbjct: 671 -----ATSSPAPSRPSSETSPPSASSTSATTTPTPAASRPRLETCPT------------- 796
Query: 466 YSGGKIAPNSSPQISFSRSSSRS 488
S G P ++ F RSS S
Sbjct: 797 -SSG*TPPTAASPARFRRSSGSS 862
>TC231933 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(65%)
Length = 619
Score = 42.0 bits (97), Expect = 7e-04
Identities = 41/134 (30%), Positives = 58/134 (42%), Gaps = 1/134 (0%)
Frame = +3
Query: 296 PSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSE 355
PSS+P S S S A A + SPSP + S ++AS PS LPP P++
Sbjct: 75 PSSIPTS---SSSSPPSSALSSASSASSPSPAAAVS---AASASPPTTLLPSLLPPPPTK 236
Query: 356 MSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSES-APVNIPNAEVT 414
S PSP P P+P SSSP S +S+ +SE ++ +
Sbjct: 237 ASRRRSSA-------PSPRSPPPPNPLSSSPTARSASPSSQPETKSECFLSADMASTSAA 395
Query: 415 YSPGHTSRQNLPPS 428
+PG +PP+
Sbjct: 396 STPGSDRIPPVPPA 437
>TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {Arabidopsis
thaliana;} , partial (68%)
Length = 1307
Score = 42.0 bits (97), Expect = 7e-04
Identities = 48/179 (26%), Positives = 71/179 (38%), Gaps = 12/179 (6%)
Frame = +2
Query: 283 SLPVVRMPSHGSSPSSLPFSRRHSWSYENCR--ASPPAVNYYSPSPTHSESHTLVSNASS 340
S P+ PS PS+ P + S + ++PP V+ P+PT++ +++
Sbjct: 500 SSPLPFNPSTPPKPSAPPTTSTASSASLRLAFPSAPPLVSAMPPTPTNAPPPPSPPPSAT 679
Query: 341 QGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLR 400
PPSS P PS S PSP +PS + S+ S S + K
Sbjct: 680 ----PPSSTSPSPSTSST---------SAAPSPPSTPSSNTPSARRTSSSTSSSPKPTSN 820
Query: 401 SESAPVN----------IPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQS 449
S P + IP + T SP + + + PST I+ TS SR L S
Sbjct: 821 PSSNPPSLN*TSRSITLIPRSSAT*SPPPSDKPSNNPSTTPEITSLTSSNPASRGLYTS 997
>TC209971 similar to UP|Q9MA24 (Q9MA24) T5E21.9, partial (73%)
Length = 1348
Score = 40.8 bits (94), Expect = 0.002
Identities = 19/39 (48%), Positives = 27/39 (68%)
Frame = +2
Query: 371 PSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIP 409
PSP+ +PSP PSSS P SL +S+T + ++S P +IP
Sbjct: 128 PSPSGAPSPPPSSSPPFLSLAWSSSETSIPTDSYPASIP 244
>TC212457 UP|Q39871 (Q39871) Maturation polypeptide, complete
Length = 1741
Score = 40.8 bits (94), Expect = 0.002
Identities = 47/145 (32%), Positives = 59/145 (40%), Gaps = 9/145 (6%)
Frame = -1
Query: 292 HGSSP----SSLPFSRRHSWSYEN-----CRASPPAVNYYSPSPTHSESHTLVSNASSQG 342
H SSP SSLP S R +S C SPP Y SP P S +L+ +S
Sbjct: 1138 HPSSPKPYSSSLPRSPRSCFSRPPPSPRLCP*SPPP--YSSPPPRSPPSFSLLPPSSPLP 965
Query: 343 YCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSE 402
Y P P PS L ++ N P P+ SP P P S+P S S S +
Sbjct: 964 YSPSPRRSPKPSL*FPLSCRSSN-----PRPS-SPHPKPCPSAPQASPTSYLSPLSPSPQ 803
Query: 403 SAPVNIPNAEVTYSPGHTSRQNLPP 427
P +P +YS H S + P
Sbjct: 802 CNPYTLP----SYSQWHLSPLSFSP 740
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 40.4 bits (93), Expect = 0.002
Identities = 41/122 (33%), Positives = 52/122 (42%), Gaps = 6/122 (4%)
Frame = +3
Query: 316 PPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNF 375
PP+ SPS T S S + + +S PPSS PS S + SP
Sbjct: 258 PPSATPPSPSATSSPSASPTAPPTSPSPSPPSS----PSPPSAPSPSSTAPSPPSASPPT 425
Query: 376 SPSPSPSSSSPIYSLGSLA--SKTLLRSESAP----VNIPNAEVTYSPGHTSRQNLPPST 429
SPSPSP S + S S A S T S ++P + P A + SP S + P T
Sbjct: 426 SPSPSPPSPASTASANSPASGSPTSRTSPTSPSPTSTSKPPAPSSSSPA*PSSKPSPSPT 605
Query: 430 PI 431
PI
Sbjct: 606 PI 611
Score = 37.4 bits (85), Expect = 0.017
Identities = 46/154 (29%), Positives = 58/154 (36%), Gaps = 4/154 (2%)
Frame = +3
Query: 248 PCVSDVSSEP--TTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRH 305
P S S+ P TP SP + SP PS P P SPSS S
Sbjct: 231 PAPSPPSTMPPSATPPSPSATSSPSASPTAPPTSPSPSPPSSPSPPSAPSPSSTAPSPPS 410
Query: 306 SWSYENCRASPPAVNYYSPSPTH-SESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNV 364
ASPP SPSP+ S + T +N+ + G + P PS S
Sbjct: 411 --------ASPPT----SPSPSPPSPASTASANSPASGSPTSRTSPTSPSPTSTSKPPAP 554
Query: 365 NFDEYY-PSPNFSPSPSPSSSSPIYSLGSLASKT 397
+ PS SPSP+P S P + + S T
Sbjct: 555 SSSSPA*PSSKPSPSPTPISPDPSPATSTPTSHT 656
Score = 33.1 bits (74), Expect = 0.32
Identities = 36/133 (27%), Positives = 53/133 (39%)
Frame = +3
Query: 345 PPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESA 404
PPS++PP + S + + +P SPSPSP SS S S +S ++
Sbjct: 243 PPSTMPPSATPPSPSATSSPSASP--TAPPTSPSPSPPSSPSPPSAPSPSSTAPSPPSAS 416
Query: 405 PVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQ 464
P P+ SP T+ N P S S T R+ S T+ K + S
Sbjct: 417 PPTSPSPSPP-SPASTASANSPAS--------GSPTSRTSPTSPSPTSTSKPPAPSSSSP 569
Query: 465 KYSGGKIAPNSSP 477
K +P+ +P
Sbjct: 570 A*PSSKPSPSPTP 608
>TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%)
Length = 537
Score = 40.4 bits (93), Expect = 0.002
Identities = 56/186 (30%), Positives = 72/186 (38%), Gaps = 4/186 (2%)
Frame = +1
Query: 251 SDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYE 310
S + + P P SP YV +P + S P V P SPSS P
Sbjct: 16 SPIYAPPIAP-SPGNHPPYVPTPPKTPSPSY-SPPNVPTPPKTPSPSSQPPFTPTPPKTP 189
Query: 311 NCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY 370
+ + PP Y P+P S S A+ P+S PP+PS +
Sbjct: 190 SPTSQPP----YIPTPPSPISQP-PSIATPPNTLSPTSQPPYPSTIP---------PSTS 327
Query: 371 PSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPN---AEVTYSPGHTSRQNLP- 426
SP++ PSPSP SS+P Y LA T S S P A T SP +S
Sbjct: 328 LSPSYPPSPSP-SSAPTYPPSYLAPATSPPSPSTSATTPTISPAATTPSPSSSSSSGASN 504
Query: 427 PSTPIR 432
+TP R
Sbjct: 505 ETTPAR 522
>TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxidase ATP26a
{Arabidopsis thaliana;} , partial (49%)
Length = 731
Score = 40.4 bits (93), Expect = 0.002
Identities = 42/166 (25%), Positives = 66/166 (39%), Gaps = 3/166 (1%)
Frame = +3
Query: 324 PSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSS 383
P P + S T S+ ++ PPSS PP PS PS +P+S
Sbjct: 183 PPPRSASSSTTASSPTA--VTPPSSSPPRPSP--------------------GPSATPTS 296
Query: 384 SSPIYSLGSLASKTLLRSESAPVNIPNAEVTYS---PGHTSRQNLPPSTPIRISRCTSET 440
+SP + S +S S+P P+ T+S P +S + PS+P + T+E
Sbjct: 297 TSPSPATPSTSSSAPRPPLSSPAPTPSPAPTFSPPPPATSSPCSAAPSSPSSSAAATAEP 476
Query: 441 ERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSS 486
S + P S+ + + + NSSP + + S S
Sbjct: 477 P-SPPPYPPTSPPPPCPSLKSPNSSPNAASPSKNSSPSVGPTPSDS 611
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.127 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,955,134
Number of Sequences: 63676
Number of extensions: 540772
Number of successful extensions: 6674
Number of sequences better than 10.0: 454
Number of HSP's better than 10.0 without gapping: 4989
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5984
length of query: 535
length of database: 12,639,632
effective HSP length: 102
effective length of query: 433
effective length of database: 6,144,680
effective search space: 2660646440
effective search space used: 2660646440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146842.6


