

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
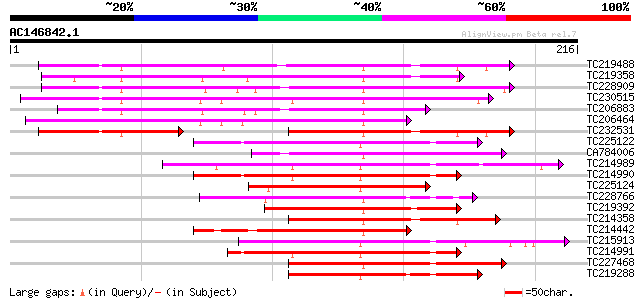
Query= AC146842.1 + phase: 0
(216 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219488 UP|Q8S900 (Q8S900) Syringolide-induced protein 1-3-1A, ... 110 4e-25
TC219358 homologue to PIR|F96527|F96527 protein F27J15.20 [impor... 94 6e-20
TC228909 similar to GB|AAS09999.1|41618996|AY519529 MYB transcri... 91 5e-19
TC230515 similar to GB|AAS09999.1|41618996|AY519529 MYB transcri... 87 8e-18
TC206883 similar to GB|AAS10003.1|41619012|AY519533 MYB transcri... 81 4e-16
TC206464 similar to GB|AAS58519.1|45357120|AY550308 MYB transcri... 80 5e-16
TC232531 UP|Q8S8Z9 (Q8S8Z9) Syringolide-induced protein 1-3-1B, ... 76 1e-14
TC225122 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1... 74 7e-14
CA784006 73 9e-14
TC214989 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1... 71 4e-13
TC214990 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1... 70 6e-13
TC225124 homologue to UP|Q8S9H7 (Q8S9H7) MYB-like transcription ... 68 4e-12
TC228766 similar to GB|AAS99672.1|46518381|BT012528 At5g56840 {A... 67 6e-12
TC219392 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1... 67 6e-12
TC214358 homologue to UP|Q84UB0 (Q84UB0) Transcription factor My... 65 2e-11
TC214442 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1... 65 2e-11
TC215913 similar to GB|AAS09986.1|41618940|AY519516 MYB transcri... 65 2e-11
TC214991 similar to GB|AAS09980.1|41618916|AY519510 MYB transcri... 65 2e-11
TC227468 similar to UP|Q8H1D0 (Q8H1D0) Transcription factor MYBS... 65 3e-11
TC219288 similar to GB|AAS99672.1|46518381|BT012528 At5g56840 {A... 64 5e-11
>TC219488 UP|Q8S900 (Q8S900) Syringolide-induced protein 1-3-1A, complete
Length = 1338
Score = 110 bits (276), Expect = 4e-25
Identities = 77/200 (38%), Positives = 112/200 (55%), Gaps = 19/200 (9%)
Frame = +2
Query: 12 EWTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIEEGLV 70
+WT +K+FE L E++ + RWE I V G+S+ EV+EHYE L+HD+ I+ G V
Sbjct: 254 QWTRYHDKLFERALLVVPEDLPD-RWEKIADQVPGKSAVEVREHYEALVHDVFEIDSGRV 430
Query: 71 DFSTNSDDFI---------------ISKASTDENKAPPTKNKTKKVVRVKHWTEEEHRLF 115
+ + DD + ++ S +N+ KK WTEEEHRLF
Sbjct: 431 EVPSYVDDSVAMPPSGGAGISTWDNANQISFGSKLKQQGENERKKGT---PWTEEEHRLF 601
Query: 116 LEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFY-ITSL 173
L G+ GKG W+ IS++ V TRT +QVASHAQK+FL Q S K +KRS+ + IT++
Sbjct: 602 LIGLSKFGKGDWRSISRNVVVTRTPTQVASHAQKYFLRQ---NSVKKERKRSSIHDITTV 772
Query: 174 KGNSKPL-LNKDNIPSPSTS 192
NS P+ +++ +P P S
Sbjct: 773 DSNSAPMPIDQTWVPPPGGS 832
>TC219358 homologue to PIR|F96527|F96527 protein F27J15.20 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(39%)
Length = 1195
Score = 93.6 bits (231), Expect = 6e-20
Identities = 67/197 (34%), Positives = 104/197 (52%), Gaps = 36/197 (18%)
Frame = +3
Query: 13 WTWDENKIFET-ILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIEEGLV 70
W++DE K FE I ++EE + +WE I V +S EVK+HY+ L+ D++ IE G +
Sbjct: 51 WSYDEEKAFENAIAMHWIEESSKEQWEKIASAVPSKSMEEVKQHYQVLVEDVSAIEAGHI 230
Query: 71 DF---------STNSDDFIISKASTDEN-----------------------KAPPTKNKT 98
F S+N D SKA++ + K +++
Sbjct: 231 SFPNYASDETTSSNKDFHGSSKATSSDKRSNCNYGSGFSGLGLDSTTHSSGKGGLSRSSE 410
Query: 99 KKVVRVKHWTEEEHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGT 157
++ + WTEEEHRLFL G++ GKG W+ IS++ V +RT +QVASHAQK+F+
Sbjct: 411 QERRKGIPWTEEEHRLFLLGLDKFGKGDWRSISRNFVISRTPTQVASHAQKYFIRL---N 581
Query: 158 SKKTYKKRSNFY-ITSL 173
S ++RS+ + ITS+
Sbjct: 582 SMNRDRRRSSIHDITSV 632
>TC228909 similar to GB|AAS09999.1|41618996|AY519529 MYB transcription factor
{Arabidopsis thaliana;} , partial (60%)
Length = 1612
Score = 90.5 bits (223), Expect = 5e-19
Identities = 67/200 (33%), Positives = 103/200 (51%), Gaps = 20/200 (10%)
Frame = +1
Query: 13 WTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIEEGLVD 71
WT ENK+FE L + ++ + RW + L+ G++ +V + Y L D+++IE G +
Sbjct: 559 WTPQENKLFENALAVFDKDTPD-RWLKVAALIPGKTVGDVIKQYRELEEDVSVIESGFIP 735
Query: 72 FS--TNSDDFIISKAS-------------TDENKAP--PTKNKTKKVVRVKHWTEEEHRL 114
T +D F + + T + A P++ + KK V WT+EEHR
Sbjct: 736 LPGYTATDSFTLEWVNNQGFGGLRQFYGVTGKRGASTRPSEQERKKGVP---WTKEEHRQ 906
Query: 115 FLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITSL 173
FL G++ +GKG W+ IS++ V TRT +QVASHAQK+F+ QL G K + + +L
Sbjct: 907 FLMGLKKYGKGDWRNISRNFVITRTPTQVASHAQKYFIRQLSGGKDKKRSSIHDITMVNL 1086
Query: 174 KGNSKPLLNKDNIP-SPSTS 192
P +N P SP S
Sbjct: 1087PEAKSPSSESNNRPYSPDHS 1146
>TC230515 similar to GB|AAS09999.1|41618996|AY519529 MYB transcription factor
{Arabidopsis thaliana;} , partial (50%)
Length = 946
Score = 86.7 bits (213), Expect = 8e-18
Identities = 65/191 (34%), Positives = 102/191 (53%), Gaps = 11/191 (5%)
Frame = +1
Query: 5 NNDGKSKEWTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLA 63
+ +G+ EWT +ENK FE+ L Y ++ + RW + ++ G++ +V + Y L D+
Sbjct: 139 SQEGQFTEWTREENKKFESALAIYDKDTPD-RWLRVAAMLPGKTVYDVIKQYRELEEDVC 315
Query: 64 LIEEGLVD---FSTNSDDF-IISKASTDENKAPPTKNKTKKVVRVKH--WTEEEHRLFLE 117
IE G + + T+S ++ D + P ++ R K WTEEEHR FL
Sbjct: 316 EIEAGRIPVPGYPTSSFTLKMVDNQCYDACRKKPATLRSSDQERKKGVPWTEEEHRRFLM 495
Query: 118 GIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITSLKGN 176
G+ +GKG W+ IS++ V T+T +QVASHAQK+++ Q K ++ S IT +
Sbjct: 496 GLLKYGKGDWRNISRNFVVTKTPTQVASHAQKYYIRQKLSGGKDNKRRPSIHDITIVNLT 675
Query: 177 S---KPLLNKD 184
S KPLL D
Sbjct: 676 SDQEKPLLLND 708
>TC206883 similar to GB|AAS10003.1|41619012|AY519533 MYB transcription factor
{Arabidopsis thaliana;} , partial (51%)
Length = 1224
Score = 80.9 bits (198), Expect = 4e-16
Identities = 58/159 (36%), Positives = 88/159 (54%), Gaps = 17/159 (10%)
Frame = +3
Query: 19 KIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIEEGLVDF----S 73
K+FE L + ++ + RW + ++ G++ +V Y+ L D++ IE GL+ S
Sbjct: 3 KLFENALAVHDKDTPD-RWHKVAEMIPGKTVVDVIRQYKELEVDVSNIEAGLIPVPGYSS 179
Query: 74 TNSDDFIISKASTDE-------NKAP----PTKNKTKKVVRVKHWTEEEHRLFLEGIEIH 122
T F + +T K P P +++ KK V WTEEEH+LFL G++ +
Sbjct: 180 TAISPFTLDWVNTPGYDGFKGCGKRPSSVRPIEHERKKGVP---WTEEEHKLFLLGLKKY 350
Query: 123 GKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKK 160
GKG W+ IS++ V TRT +QVASHAQK+F+ QL G K
Sbjct: 351 GKGDWRNISRNFVITRTPTQVASHAQKYFIRQLSGGKDK 467
>TC206464 similar to GB|AAS58519.1|45357120|AY550308 MYB transcription factor
{Arabidopsis thaliana;} , partial (61%)
Length = 1156
Score = 80.5 bits (197), Expect = 5e-16
Identities = 50/154 (32%), Positives = 87/154 (56%), Gaps = 7/154 (4%)
Frame = +3
Query: 7 DGKSKEWTWDENKIFETILFEYLEEVQEGRWENIGLVCGRSSTEVKEHYETLLHDLALIE 66
+ S EWT ++NK FE+ L Y + + ++ ++ G++ +V + Y L D++ IE
Sbjct: 582 ESHSTEWTREDNKKFESALAIYDNDTPDRWFKVAAMIPGKTVFDVIKQYRELEEDVSEIE 761
Query: 67 EGLVD---FSTNSDDF-IISKASTD--ENKAPPTKNKTKKVVRVKHWTEEEHRLFLEGIE 120
G V + +S F ++ + D + P + ++ + WTE+EHR FL G+
Sbjct: 762 AGRVPIPGYLASSFTFELVDNHNYDGCRRRLAPVRGSDQERKKGVPWTEDEHRRFLMGLL 941
Query: 121 IHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQ 153
+GKG W+ IS++ V T+T +QVASHAQK+++ Q
Sbjct: 942 KYGKGDWRNISRNFVVTKTPTQVASHAQKYYIRQ 1043
>TC232531 UP|Q8S8Z9 (Q8S8Z9) Syringolide-induced protein 1-3-1B, partial
(98%)
Length = 1225
Score = 76.3 bits (186), Expect = 1e-14
Identities = 45/89 (50%), Positives = 62/89 (69%), Gaps = 3/89 (3%)
Frame = +2
Query: 107 WTEEEHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKR 165
WTEEEHRLFL G+ GKG W+ IS++ V TRT +QVASHAQK+FL Q S K +KR
Sbjct: 557 WTEEEHRLFLIGLSKFGKGDWRSISRNVVVTRTPTQVASHAQKYFLRQ---NSVKKERKR 727
Query: 166 SNFY-ITSLKGNSKPL-LNKDNIPSPSTS 192
S+ + IT++ NS P+ ++++ +P P S
Sbjct: 728 SSIHDITTVDSNSVPVPIDQNWVPPPGGS 814
Score = 45.4 bits (106), Expect = 2e-05
Identities = 24/56 (42%), Positives = 36/56 (63%), Gaps = 1/56 (1%)
Frame = +3
Query: 12 EWTWDENKIFETILFEYLEEVQEGRWENIG-LVCGRSSTEVKEHYETLLHDLALIE 66
+WT +K+FE L E++ + RWE I V G+S+ EV+EHYE L+HD+ I+
Sbjct: 234 QWTRYHDKLFERALLVVPEDLPD-RWEKIADQVPGKSAVEVREHYEALVHDVFEID 398
>TC225122 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1, partial
(40%)
Length = 1301
Score = 73.6 bits (179), Expect = 7e-14
Identities = 45/111 (40%), Positives = 67/111 (59%), Gaps = 1/111 (0%)
Frame = +1
Query: 71 DFSTNSDDFIISKASTDENKAPPTKNKTKKVVRVKHWTEEEHRLFLEGIEIHGKGKWKLI 130
D + DD AS D+ AP K + R WTEEEH+LFL G++ GKG W+ I
Sbjct: 208 DAANAKDDVAAGYASADD-AAPINSGKNRDRKRGIPWTEEEHKLFLVGLQKVGKGDWRGI 384
Query: 131 SQ-HVRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITSLKGNSKPL 180
S+ +V+TRT +QVASHAQK+FL + + ++ ++ S F IT+ ++ P+
Sbjct: 385 SRNYVKTRTPTQVASHAQKYFLRRTNLNRRR--RRSSLFDITTDSVSTTPM 531
>CA784006
Length = 409
Score = 73.2 bits (178), Expect = 9e-14
Identities = 42/98 (42%), Positives = 58/98 (58%), Gaps = 1/98 (1%)
Frame = +3
Query: 93 PTKNKTKKVVRVKHWTEEEHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFL 151
PT+ + KK V WTEEEHR FL G++ +GKG W+ IS++ V TRT +QVASHAQK+F+
Sbjct: 78 PTEQERKKGVP---WTEEEHRQFLLGLKKYGKGDWRNISRNFVTTRTPTQVASHAQKYFI 248
Query: 152 HQLDGTSKKTYKKRSNFYITSLKGNSKPLLNKDNIPSP 189
QL G K + + +L+ P + SP
Sbjct: 249 RQLTGGKDKRRSSIHDITMVNLQETKSPSSDTTKPSSP 362
>TC214989 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1, partial
(42%)
Length = 1511
Score = 70.9 bits (172), Expect = 4e-13
Identities = 51/161 (31%), Positives = 93/161 (57%), Gaps = 8/161 (4%)
Frame = +3
Query: 59 LHDLALIEEGLVDFSTNSD-DFIISKASTDENKAPPTKNKTKKVVRVKH--WTEEEHRLF 115
+++L+ E L +TN++ D + + ++ ++ AP + ++ R + WTEEEH+LF
Sbjct: 207 MNNLSQYEHPLDATTTNNNKDAVAAGYASADDAAPQNSGRHRERERKRGVPWTEEEHKLF 386
Query: 116 LEGIEIHGKGKWKLISQ-HVRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITSLK 174
L G++ GKG W+ IS+ +V+TRT +QVASHAQK+FL + + ++ ++ S F IT+
Sbjct: 387 LVGLQKVGKGDWRGISKNYVKTRTPTQVASHAQKYFLRRSNLNRRR--RRSSLFDITTDT 560
Query: 175 GNSKPLLNKDNIPSPSTSWDGNFHPLL----YKDNYVPALP 211
++ P + ++ + + T P+ K N PA+P
Sbjct: 561 VSAIP-MEEEQVQNQDTLCHSQQQPVFPAETSKINGFPAMP 680
>TC214990 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1, partial
(59%)
Length = 1443
Score = 70.5 bits (171), Expect = 6e-13
Identities = 44/105 (41%), Positives = 65/105 (61%), Gaps = 3/105 (2%)
Frame = +3
Query: 71 DFSTNSDDFIISKASTDENKAPPTKNKTKKVVRVKH--WTEEEHRLFLEGIEIHGKGKWK 128
D S N D AS D+ AP + ++ R + WTEEEH+LFL G++ GKG W+
Sbjct: 255 DGSNNKDALAAGYASADD-AAPQNSGRLRERERKRGVPWTEEEHKLFLVGLQKVGKGDWR 431
Query: 129 LISQ-HVRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITS 172
IS+ +V+TRT +QVASHAQK+FL + + ++ ++ S F IT+
Sbjct: 432 GISKNYVKTRTPTQVASHAQKYFLRRSNLNRRR--RRSSLFDITT 560
>TC225124 homologue to UP|Q8S9H7 (Q8S9H7) MYB-like transcription factor
DIVARICATA, partial (27%)
Length = 483
Score = 67.8 bits (164), Expect = 4e-12
Identities = 36/71 (50%), Positives = 51/71 (71%), Gaps = 2/71 (2%)
Frame = +2
Query: 92 PPTKNK-TKKVVRVKHWTEEEHRLFLEGIEIHGKGKWKLISQ-HVRTRTASQVASHAQKH 149
PP ++ ++K +V H EEEH+LFL G++ +GKG W+ IS+ +V TRT +QVASHAQK+
Sbjct: 59 PPLEDPLSRKGRKVCHGXEEEHKLFLLGLKKYGKGDWRNISRNYVITRTPTQVASHAQKY 238
Query: 150 FLHQLDGTSKK 160
F+ QL G K
Sbjct: 239 FIRQLSGGKDK 271
>TC228766 similar to GB|AAS99672.1|46518381|BT012528 At5g56840 {Arabidopsis
thaliana;} , partial (54%)
Length = 1220
Score = 67.0 bits (162), Expect = 6e-12
Identities = 47/115 (40%), Positives = 69/115 (59%), Gaps = 9/115 (7%)
Frame = +3
Query: 73 STNSDDFIISKASTDENKAPPTKN--------KTKKVVRVKHWTEEEHRLFLEGIEIHGK 124
S++S F S+ + DEN + + ++ + WTEEEHR+FL G+E GK
Sbjct: 285 SSSSSSFSSSRLTIDENSDRTSFGYLSDGLLARAQERKKGVPWTEEEHRIFLVGLEKLGK 464
Query: 125 GKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKRSNFYITSLKGNSK 178
G W+ IS++ V TRT +QVASHAQK+FL +L KK K+RS+ + L G++K
Sbjct: 465 GDWRGISRNFVTTRTPTQVASHAQKYFL-RLATMDKK--KRRSSLF--DLVGSNK 614
>TC219392 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1, partial
(28%)
Length = 918
Score = 67.0 bits (162), Expect = 6e-12
Identities = 38/76 (50%), Positives = 53/76 (69%), Gaps = 1/76 (1%)
Frame = +3
Query: 98 TKKVVRVKHWTEEEHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDG 156
T++ R WTEEEHRLFL G++ GKG W+ IS++ V+TRT +QVASHAQK+FL +
Sbjct: 366 TRERKRGVPWTEEEHRLFLLGLQNIGKGDWRGISRNFVKTRTPTQVASHAQKYFLRR--H 539
Query: 157 TSKKTYKKRSNFYITS 172
T + ++ S F IT+
Sbjct: 540 TQNRRRRRSSLFDITT 587
>TC214358 homologue to UP|Q84UB0 (Q84UB0) Transcription factor Myb1, partial
(24%)
Length = 940
Score = 65.5 bits (158), Expect = 2e-11
Identities = 36/84 (42%), Positives = 55/84 (64%), Gaps = 3/84 (3%)
Frame = +2
Query: 107 WTEEEHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKR 165
WTEEEHRLFL G+ GKG W+ IS++ V+TRT +QVASHAQK+FL + ++ ++R
Sbjct: 11 WTEEEHRLFLLGLHKVGKGDWRGISRNFVKTRTPTQVASHAQKYFLRR---HNQNRRRRR 181
Query: 166 SNFY--ITSLKGNSKPLLNKDNIP 187
S+ + T S ++ ++ +P
Sbjct: 182 SSLFDITTDTVMESSTIMEEEQVP 253
>TC214442 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1, partial
(30%)
Length = 1102
Score = 65.5 bits (158), Expect = 2e-11
Identities = 41/84 (48%), Positives = 53/84 (62%), Gaps = 1/84 (1%)
Frame = +2
Query: 71 DFSTNSDDFIISKASTDENKAPPTKNKTKKVVRVKHWTEEEHRLFLEGIEIHGKGKWKLI 130
D N+DD AS D A +T++ R WTEEEHRLFL G+ GKG W+ I
Sbjct: 377 DAEFNADD--AGYASDDVVHA---SGRTRERKRGVPWTEEEHRLFLLGLHKVGKGDWRGI 541
Query: 131 SQH-VRTRTASQVASHAQKHFLHQ 153
S++ V+TRT +QVASHAQK+FL +
Sbjct: 542 SRNFVKTRTPTQVASHAQKYFLRR 613
>TC215913 similar to GB|AAS09986.1|41618940|AY519516 MYB transcription factor
{Arabidopsis thaliana;} , partial (66%)
Length = 1545
Score = 65.5 bits (158), Expect = 2e-11
Identities = 52/165 (31%), Positives = 79/165 (47%), Gaps = 39/165 (23%)
Frame = +2
Query: 88 ENKAPPTKNKTKKVVRVKHWTEEEHRLFLEGIEIHGKGKWKLISQ-HVRTRTASQVASHA 146
E+ P + + +++ + WTEEEHR+FL G++ GKG W+ I++ +V +RT +QVASHA
Sbjct: 452 EDFVPGSSSSSRERKKGVPWTEEEHRMFLLGLQKLGKGDWRGIARNYVISRTPTQVASHA 631
Query: 147 QKHFLHQLDGTSKKTYKKRSNFYITS-------------LKGNSKPLLNKDNIPSP---- 189
QK+F+ Q + + +K ++ S F I + L N P + N P P
Sbjct: 632 QKYFIRQSNVSRRK--RRSSLFDIVADEAADTAMVQQDFLSANELPTETEGNNPLPAPPP 805
Query: 190 ----------STSWDG--------NFH---PLLYKDNYVPALPSP 213
+ S DG N H P+LY Y P P P
Sbjct: 806 LDEECESMDSTNSNDGEPAPSKPENTHPSYPMLYPAYYSPVFPFP 940
>TC214991 similar to GB|AAS09980.1|41618916|AY519510 MYB transcription factor
{Arabidopsis thaliana;} , partial (33%)
Length = 677
Score = 65.1 bits (157), Expect = 2e-11
Identities = 40/92 (43%), Positives = 61/92 (65%), Gaps = 3/92 (3%)
Frame = +2
Query: 84 ASTDENKAPPTKNKTKKVVRVKH--WTEEEHRLFLEGIEIHGKGKWKLISQ-HVRTRTAS 140
AS D+ AP + ++ R + WTEEEH+LFL G++ GKG W+ IS+ +V+TRT +
Sbjct: 20 ASADD-AAPQNSGRHRERERKRGVPWTEEEHKLFLVGLQKVGKGDWRGISKNYVKTRTPT 196
Query: 141 QVASHAQKHFLHQLDGTSKKTYKKRSNFYITS 172
QVASHAQK+FL + + ++ ++ S F IT+
Sbjct: 197 QVASHAQKYFLRRSNLNRRR--RRSSLFDITT 286
>TC227468 similar to UP|Q8H1D0 (Q8H1D0) Transcription factor MYBS3, partial
(56%)
Length = 1404
Score = 64.7 bits (156), Expect = 3e-11
Identities = 35/84 (41%), Positives = 55/84 (64%), Gaps = 1/84 (1%)
Frame = +3
Query: 107 WTEEEHRLFLEGIEIHGKGKWKLISQH-VRTRTASQVASHAQKHFLHQLDGTSKKTYKKR 165
WTEEEHRLFL G++ GKG W+ I+++ V +RT +QVASHAQK+F+ Q T +K ++
Sbjct: 399 WTEEEHRLFLIGLQKLGKGDWRGIARNFVVSRTPTQVASHAQKYFIRQSHATRRK--RRS 572
Query: 166 SNFYITSLKGNSKPLLNKDNIPSP 189
S F + + +P + ++ + P
Sbjct: 573 SLFDMVPDMSSDQPSVPEEQVLLP 644
>TC219288 similar to GB|AAS99672.1|46518381|BT012528 At5g56840 {Arabidopsis
thaliana;} , partial (41%)
Length = 720
Score = 63.9 bits (154), Expect = 5e-11
Identities = 37/75 (49%), Positives = 52/75 (69%), Gaps = 1/75 (1%)
Frame = +2
Query: 107 WTEEEHRLFLEGIEIHGKGKWKLISQ-HVRTRTASQVASHAQKHFLHQLDGTSKKTYKKR 165
WTEEEHR FL G+E GKG W+ IS+ +V +RT +QVASHAQK+F+ +L +KK K+R
Sbjct: 314 WTEEEHRTFLVGLEKLGKGDWRGISRNYVTSRTPTQVASHAQKYFI-RLATMNKK--KRR 484
Query: 166 SNFYITSLKGNSKPL 180
S+ + G + P+
Sbjct: 485 SSLFDMVGNGITNPI 529
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.131 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,118,016
Number of Sequences: 63676
Number of extensions: 157839
Number of successful extensions: 780
Number of sequences better than 10.0: 130
Number of HSP's better than 10.0 without gapping: 735
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 739
length of query: 216
length of database: 12,639,632
effective HSP length: 93
effective length of query: 123
effective length of database: 6,717,764
effective search space: 826284972
effective search space used: 826284972
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146842.1


