

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146819.11 - phase: 0
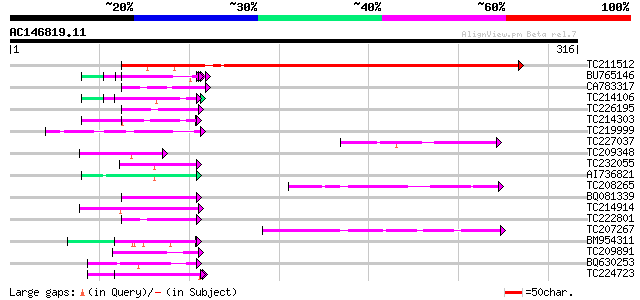
(316 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211512 similar to UP|Q91W51 (Q91W51) WASP family 1, partial (4%) 357 5e-99
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 48 7e-06
CA783317 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 45 4e-05
TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 45 6e-05
TC226195 weakly similar to UP|Q95JD0 (Q95JD0) Basic proline-rich... 44 1e-04
TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, pa... 44 1e-04
TC219999 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (5%) 44 1e-04
TC227037 homologue to UP|MSI4_ARATH (O22607) WD-40 repeat protei... 44 1e-04
TC209348 similar to UP|Q9LSD7 (Q9LSD7) Arabidopsis thaliana geno... 43 2e-04
TC232055 similar to UP|O49524 (O49524) Pherophorin - like protei... 43 2e-04
AI736821 weakly similar to GP|11036868|gb| PxORF73 peptide {Plut... 43 2e-04
TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%) 43 2e-04
BQ081339 similar to GP|27817931|db P0453E05.6 {Oryza sativa (jap... 43 2e-04
TC214914 homologue to UP|Q8LK71 (Q8LK71) Extensin-like protein, ... 43 2e-04
TC222801 weakly similar to UP|O23370 (O23370) Cell wall protein ... 43 2e-04
TC207267 similar to UP|Q8GTM1 (Q8GTM1) WD40, partial (53%) 42 3e-04
BM954311 weakly similar to GP|21322711|e pherophorin-dz1 protein... 42 3e-04
TC209891 weakly similar to UP|Q9LW52 (Q9LW52) Arabidopsis thalia... 42 3e-04
BQ630253 42 4e-04
TC224723 hydroxyproline-rich glycoprotein 42 4e-04
>TC211512 similar to UP|Q91W51 (Q91W51) WASP family 1, partial (4%)
Length = 801
Score = 357 bits (915), Expect = 5e-99
Identities = 182/235 (77%), Positives = 195/235 (82%), Gaps = 11/235 (4%)
Frame = +3
Query: 63 PPKNPNPPSPPLQ------DDSPLVGPPPPPPHH-----EADDDDGEMIGPPPPPPGSNF 111
PP + NP S DD P+ G H + +DDD EMIGPPPPPP +
Sbjct: 39 PPPSQNPYSSSSAAAARSXDDGPMXGNRXAASAHSWPTKKKNDDDAEMIGPPPPPPIA-- 212
Query: 112 NDSEDEDNDSDQDELGNRFRIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMY 171
SE ED DSDQDE+G RFRIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMY
Sbjct: 213 --SESED-DSDQDEVGTRFRIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMY 383
Query: 172 DFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKG 231
DFQGMN+RL+SFRQ+EP EGHQ+RNLSWSPTADR+LCVTGSAQAKIYDRDGLTLGEFVKG
Sbjct: 384 DFQGMNARLESFRQLEPFEGHQVRNLSWSPTADRYLCVTGSAQAKIYDRDGLTLGEFVKG 563
Query: 232 DMYIRDLKNTKGHITGLTCGEWHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWK 286
DMYIRDLKNTKGHI+GLTCGEWHPKTKETILTSSEDGSLRIWDVNDF +QKQV K
Sbjct: 564 DMYIRDLKNTKGHISGLTCGEWHPKTKETILTSSEDGSLRIWDVNDFKSQKQVIK 728
Score = 38.1 bits (87), Expect = 0.005
Identities = 20/55 (36%), Positives = 24/55 (43%)
Frame = +1
Query: 54 SKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPG 108
S++ L+S+RPPK P PP PP PPP PPPP G
Sbjct: 19 SRDWLNSLRPPKTPTPP------------PPLPPPDXXTTAQWXGTAXPPPPTAG 147
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 47.8 bits (112), Expect = 7e-06
Identities = 23/46 (50%), Positives = 23/46 (50%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPG 108
PP P PPSPPL P PPPPPP PPPPPPG
Sbjct: 125 PPPPPPPPSPPLPPPPPPPPPPPPPPPPP----------PPPPPPG 18
Score = 47.0 bits (110), Expect = 1e-05
Identities = 21/45 (46%), Positives = 22/45 (48%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP SP PPPPPP + PPPPPP
Sbjct: 200 PPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPSPPLPPPPPPPP 66
Score = 46.2 bits (108), Expect = 2e-05
Identities = 21/45 (46%), Positives = 21/45 (46%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PPL P PPPPPP PPPPPP
Sbjct: 190 PPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPP 56
Score = 45.1 bits (105), Expect = 4e-05
Identities = 24/67 (35%), Positives = 26/67 (37%)
Frame = -2
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMI 100
P +P P K+ PP P PP PP P PPPPPP
Sbjct: 262 PPPSPPPPXXSLPPKKPPPPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPSPP 83
Query: 101 GPPPPPP 107
PPPPPP
Sbjct: 82 PPPPPPP 62
Score = 45.1 bits (105), Expect = 4e-05
Identities = 23/46 (50%), Positives = 23/46 (50%), Gaps = 1/46 (2%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDD-SPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PPL SPL PPPPPP PPPPPP
Sbjct: 276 PPPPPPPPLPPLPXSLSPLKNPPPPPP-------------PPPPPP 178
Score = 44.7 bits (104), Expect = 6e-05
Identities = 22/50 (44%), Positives = 24/50 (48%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFN 112
PP P PP PP PL PPPPPP PPPPPP +N +
Sbjct: 153 PPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPP-PPPPPPPRANLS 7
Score = 44.7 bits (104), Expect = 6e-05
Identities = 20/45 (44%), Positives = 21/45 (46%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP + PPPPPP
Sbjct: 213 PPPPPPPPPPPPPPPPPPPFPPPPPPPPPPPPPPPPPLPPPPPPP 79
Score = 43.5 bits (101), Expect = 1e-04
Identities = 22/48 (45%), Positives = 23/48 (47%)
Frame = -3
Query: 60 SIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
S+ P KNP PP PP P PPPPPP PPPPPP
Sbjct: 234 SLSPLKNPPPPPPP----PPPPPPPPPPPPFPPPPPPPPPPPPPPPPP 103
Score = 43.5 bits (101), Expect = 1e-04
Identities = 21/45 (46%), Positives = 21/45 (46%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP SP PPPPPP PPPPPP
Sbjct: 366 PPPPPPPPPPPPPPPSPPPPPPPPPP-------------PPPPPP 271
Score = 42.7 bits (99), Expect = 2e-04
Identities = 21/45 (46%), Positives = 21/45 (46%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP SPL PPPP P PPPPPP
Sbjct: 373 PPPPPPPPPPPPPPPSPLPPPPPPXP-------------PPPPPP 278
Score = 42.7 bits (99), Expect = 2e-04
Identities = 24/55 (43%), Positives = 24/55 (43%)
Frame = -1
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P L S PP P PP PP PL PPPPPP PPPPPP
Sbjct: 395 PPPSSLPSPPPPPPPPPPPPP-----PLPPPPPPPP------------XPPPPPP 282
Score = 42.0 bits (97), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 353 PPPPPPPPLPPPPPPPPXPPPPPPPP-------------PPPPPP 258
Score = 41.6 bits (96), Expect = 5e-04
Identities = 20/45 (44%), Positives = 21/45 (46%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP + PPPPPP
Sbjct: 318 PPPPPPPPPPP----PPPPPPPPPPPLPPLPXSLSPLKNPPPPPP 196
Score = 41.6 bits (96), Expect = 5e-04
Identities = 20/47 (42%), Positives = 22/47 (46%), Gaps = 2/47 (4%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEM--IGPPPPPP 107
PP +P PP PP P PPPPPP + PPPPPP
Sbjct: 337 PPPSPLPPPPPPXPPPPPPPPPPPPPPPSPPPPXXSLPPKKPPPPPP 197
Score = 41.2 bits (95), Expect = 6e-04
Identities = 22/55 (40%), Positives = 23/55 (41%)
Frame = -3
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P L + PP P PP PP P PPPPPP PPPPPP
Sbjct: 399 PPSSLLPPLPPPPPPPPPPPP---PPPPPSPPPPPP-------------PPPPPP 283
Score = 41.2 bits (95), Expect = 6e-04
Identities = 21/47 (44%), Positives = 21/47 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGS 109
PP P PPSP P PPPPPP PPPPPP S
Sbjct: 352 PPPPPPPPSPLPPPPPPXPPPPPPPP-------------PPPPPPPS 251
Score = 40.4 bits (93), Expect = 0.001
Identities = 21/52 (40%), Positives = 22/52 (41%), Gaps = 7/52 (13%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGP-------PPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PPL S + P PPPPP PPPPPP
Sbjct: 287 PPPPPPPPPPPLSPPSLXLSPP*KTPPPPPPPPPPPPPPPPPPPSPPPPPPP 132
Score = 40.0 bits (92), Expect = 0.001
Identities = 20/48 (41%), Positives = 22/48 (45%)
Frame = -3
Query: 60 SIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
S+ PP P PP PP PPPPPP + PPPPPP
Sbjct: 390 SLLPPLPPPPPPPP---------PPPPPPPPPSPPPPPPPPPPPPPPP 274
Score = 39.7 bits (91), Expect = 0.002
Identities = 21/47 (44%), Positives = 21/47 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGS 109
P K P PP PP P PPPPPP PPPPPP S
Sbjct: 224 P*KTPPPPPPP-----PPPPPPPPPPPPSPPPPPPPPPPPPPPPPPS 99
Score = 38.9 bits (89), Expect = 0.003
Identities = 20/48 (41%), Positives = 20/48 (41%), Gaps = 3/48 (6%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADD---DDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 335 PPLPPPPPPPPXPPPPPPPPPPPPPPPLSPPSLXLSPP*KTPPPPPPP 192
Score = 35.8 bits (81), Expect = 0.026
Identities = 17/38 (44%), Positives = 17/38 (44%)
Frame = -3
Query: 70 PSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P PP PL PPPPPP PPPPPP
Sbjct: 405 PPPPSSLLPPLPPPPPPPPPPPPPPPPPSPPPPPPPPP 292
Score = 35.0 bits (79), Expect = 0.045
Identities = 19/45 (42%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP PPSPP PPPPPP + PPPPPP
Sbjct: 403 PPPLLPPPSPP---------PPPPPPPPPPPPPPSPL--PPPPPP 302
Score = 33.9 bits (76), Expect = 0.099
Identities = 22/67 (32%), Positives = 24/67 (34%)
Frame = -1
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMI 100
P+S P P + PP P P PP PPPPPP
Sbjct: 389 PSSLPSPPPPPPPPPPPPPPLPPPPPPPPXPPP---------PPPPPP------------ 273
Query: 101 GPPPPPP 107
PPPPPP
Sbjct: 272 -PPPPPP 255
>CA783317 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (23%)
Length = 159
Score = 45.1 bits (105), Expect = 4e-05
Identities = 24/50 (48%), Positives = 25/50 (50%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFN 112
PP P PP PP SPL PPPPPP PPPPPPG F+
Sbjct: 159 PPPLPPPPPPP---PSPLXXPPPPPP--PPPPPPSPPPPPPPPPPGLLFS 25
Score = 36.6 bits (83), Expect = 0.015
Identities = 22/59 (37%), Positives = 22/59 (37%)
Frame = -2
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNF 111
P L S PP P PP PP PPPPPP PPPPP F
Sbjct: 137 PPPLLLXSXXPPPPPPPPPPP---------PPPPPPX-------------PPPPPACFF 27
Score = 34.7 bits (78), Expect = 0.058
Identities = 19/43 (44%), Positives = 20/43 (46%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPP 105
PP P PPSPP PPPPPP G + PPPP
Sbjct: 96 PPPPPPPPSPP---------PPPPPPPPGLLFSGGSL--PPPP 1
>TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (22%)
Length = 1296
Score = 44.7 bits (104), Expect = 6e-05
Identities = 22/52 (42%), Positives = 26/52 (49%), Gaps = 3/52 (5%)
Frame = +1
Query: 59 HSIRPPKNPNPPSPPLQDDSPL---VGPPPPPPHHEADDDDGEMIGPPPPPP 107
HS PP +PP PP P+ + PPPPPP H + PPPPPP
Sbjct: 220 HSPPPPPPHSPPPPPPSPPPPVYPYLSPPPPPPVHSPPPP---VYSPPPPPP 366
Score = 43.5 bits (101), Expect = 1e-04
Identities = 22/69 (31%), Positives = 27/69 (38%)
Frame = +1
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMI 100
P +P P + +R P P+PP P P V PPPP +
Sbjct: 43 PVYSPPPPPPSPPPPSPTYCVRSPPPPSPPPPSPPPPPPPVFSPPPPVQYYYSSPPPPQH 222
Query: 101 GPPPPPPGS 109
PPPPPP S
Sbjct: 223 SPPPPPPHS 249
Score = 42.7 bits (99), Expect = 2e-04
Identities = 23/55 (41%), Positives = 25/55 (44%)
Frame = +1
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P +HS PP PP PP P + PPPPPP I PPPPPP
Sbjct: 304 PPPPPVHSPPPPVYSPPPPPP--SPPPCIEPPPPPP---------PCIEPPPPPP 435
Score = 40.4 bits (93), Expect = 0.001
Identities = 22/68 (32%), Positives = 30/68 (43%), Gaps = 3/68 (4%)
Frame = +1
Query: 41 PTSNPKTTSDDFPSKECLHSI--RPPKNPNPPSPPLQDDS-PLVGPPPPPPHHEADDDDG 97
P +P + P H PP +P+PP PP+Q +S P PPPP P + +
Sbjct: 427 PPPSPPPCEEHSPPPPSPHPAPYHPPPSPSPPPPPVQYNSPPPPSPPPPTPVYHYNSPPP 606
Query: 98 EMIGPPPP 105
PP P
Sbjct: 607 PSFPPPTP 630
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/44 (43%), Positives = 21/44 (47%)
Frame = +1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPP 106
PP +P PPSPP P V PPPPP + PPPP
Sbjct: 7 PPPSPPPPSPP-----PPVYSPPPPPPSPPPPSPTYCVRSPPPP 123
Score = 38.5 bits (88), Expect = 0.004
Identities = 24/78 (30%), Positives = 30/78 (37%), Gaps = 6/78 (7%)
Frame = +1
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNP-----PSPPLQDDSPLVGPPPP-PPHHEADD 94
P S P P C+ PP +P P P PP +P PP P PP
Sbjct: 361 PPSPPPCIEPPPPPPPCIEPPPPPPSPPPCEEHSPPPPSPHPAPYHPPPSPSPPPPPVQY 540
Query: 95 DDGEMIGPPPPPPGSNFN 112
+ PPPP P ++N
Sbjct: 541 NSPPPPSPPPPTPVYHYN 594
Score = 37.7 bits (86), Expect = 0.007
Identities = 22/56 (39%), Positives = 23/56 (40%), Gaps = 11/56 (19%)
Frame = +1
Query: 63 PPKNPNP------PSPPLQ-----DDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P P P PP+Q P PPPPPPH PPPPPP
Sbjct: 133 PPSPPPPPPPVFSPPPPVQYYYSSPPPPQHSPPPPPPH-----------SPPPPPP 267
Score = 36.6 bits (83), Expect = 0.015
Identities = 17/45 (37%), Positives = 21/45 (45%)
Frame = +1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +P P P P + PPPPPP ++ PPPP P
Sbjct: 358 PPPSPPPCIEPPPPPPPCIEPPPPPPSPPPCEEH----SPPPPSP 480
Score = 34.3 bits (77), Expect = 0.076
Identities = 25/83 (30%), Positives = 34/83 (40%), Gaps = 3/83 (3%)
Frame = +1
Query: 28 PLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGP-PPP 86
P + H A P+ +P P ++ PP +P PP+P +SP PPP
Sbjct: 466 PPPSPHPAPYHPPPSPSP-------PPPPVQYNSPPPPSPPPPTPVYHYNSPPPPSFPPP 624
Query: 87 PPHHEADDDD--GEMIGPPPPPP 107
P +E G PPPPP
Sbjct: 625 TPVYEGPLPPVIGVSYASPPPPP 693
Score = 33.5 bits (75), Expect = 0.13
Identities = 21/68 (30%), Positives = 25/68 (35%), Gaps = 1/68 (1%)
Frame = +1
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGP-PPPPPHHEADDDDGEM 99
P P P + +S PP +PP PP P PPPP
Sbjct: 142 PPPPPPPVFSPPPPVQYYYSSPPPPQHSPPPPP---------PHSPPPPPPSPPPPVYPY 294
Query: 100 IGPPPPPP 107
+ PPPPPP
Sbjct: 295 LSPPPPPP 318
Score = 32.7 bits (73), Expect = 0.22
Identities = 17/42 (40%), Positives = 19/42 (44%)
Frame = +1
Query: 66 NPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
+P PPSPP PP PPP + PPPPPP
Sbjct: 1 SPPPPSPP---------PPSPPP---------PVYSPPPPPP 72
>TC226195 weakly similar to UP|Q95JD0 (Q95JD0) Basic proline-rich protein,
partial (17%)
Length = 654
Score = 43.9 bits (102), Expect = 1e-04
Identities = 22/47 (46%), Positives = 24/47 (50%), Gaps = 1/47 (2%)
Frame = +3
Query: 63 PPKNPNPPSPPLQ-DDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPG 108
P +P PP+PPL SP PPP PPHH P PPPPG
Sbjct: 117 PHHHPGPPAPPLHAHPSP---PPPGPPHHHHGPPGPPHAYPTPPPPG 248
Score = 38.1 bits (87), Expect = 0.005
Identities = 30/99 (30%), Positives = 35/99 (35%), Gaps = 15/99 (15%)
Frame = +3
Query: 25 SQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPP----SPPLQDDSPL 80
S T L +H P P P LH+ P P PP PP +
Sbjct: 63 SSTTLSNMHNPP----PPGGPGPHHHPGPPAPPLHAHPSPPPPGPPHHHHGPPGPPHAYP 230
Query: 81 VGPPPPPPHHEADDDDG---EMIGPP--------PPPPG 108
PPP PPHH + + GPP PPPPG
Sbjct: 231 TPPPPGPPHHHPGPPEPGFHQHHGPPGPPYAHPTPPPPG 347
Score = 33.5 bits (75), Expect = 0.13
Identities = 18/45 (40%), Positives = 19/45 (42%)
Frame = +3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P +P PP P GPP PP H G GPPPP P
Sbjct: 252 PHHHPGPPEPGFHQHH---GPPGPPYAHPTPPPPGPP-GPPPPGP 374
Score = 30.8 bits (68), Expect = 0.84
Identities = 14/26 (53%), Positives = 15/26 (56%), Gaps = 1/26 (3%)
Frame = +3
Query: 67 PNPPSPPLQDDSPLVGPPPP-PPHHE 91
P PP PP GPPPP PPHH+
Sbjct: 333 PPPPGPP--------GPPPPGPPHHQ 386
>TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, partial (18%)
Length = 1014
Score = 43.9 bits (102), Expect = 1e-04
Identities = 20/45 (44%), Positives = 24/45 (52%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP + PP PP+ P + PPPPPP H + PPPPPP
Sbjct: 80 PPPHSPPPPPPVY---PYLSPPPPPPVHSPPPP---VYSPPPPPP 196
Score = 41.6 bits (96), Expect = 5e-04
Identities = 23/68 (33%), Positives = 29/68 (41%), Gaps = 2/68 (2%)
Frame = +2
Query: 41 PTSNPKTTSDDFPSKECLHSI--RPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGE 98
P P + P HS PP +P+PP PP+Q +SP PPPP +
Sbjct: 227 PPPPPPPCEEHSPPPPSPHSAPYHPPPSPSPPPPPIQYNSPPPPSPPPPTPVYHYNSPPP 406
Query: 99 MIGPPPPP 106
PPP P
Sbjct: 407 PSSPPPAP 430
Score = 39.7 bits (91), Expect = 0.002
Identities = 19/45 (42%), Positives = 21/45 (46%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +PP+PP P P PPPPH PPPPPP
Sbjct: 11 PPPVYSPPAPPPTYSPPPPPPSPPPPH-----------SPPPPPP 112
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/61 (39%), Positives = 27/61 (43%), Gaps = 6/61 (9%)
Frame = +2
Query: 53 PSKECLHSIRPPK------NPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPP 106
PS HS PP +P PP P P+ PPPPPP + E PPPPP
Sbjct: 71 PSPPPPHSPPPPPPVYPYLSPPPPPPVHSPPPPVYSPPPPPP---SPPPCIESPPPPPPP 241
Query: 107 P 107
P
Sbjct: 242 P 244
Score = 39.3 bits (90), Expect = 0.002
Identities = 18/45 (40%), Positives = 21/45 (46%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +PP PP P PPPPP + + PPPPPP
Sbjct: 38 PPPTYSPPPPPPSPPPPHSPPPPPPVY--------PYLSPPPPPP 148
Score = 39.3 bits (90), Expect = 0.002
Identities = 26/60 (43%), Positives = 28/60 (46%), Gaps = 3/60 (5%)
Frame = +2
Query: 53 PSKECLHSIRPP---KNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGS 109
P +HS PP P PPSPP +SP PPPPPP E PPPP P S
Sbjct: 134 PPPPPVHSPPPPVYSPPPPPPSPPPCIESP-PPPPPPPPCEEH--------SPPPPSPHS 286
Score = 36.6 bits (83), Expect = 0.015
Identities = 23/67 (34%), Positives = 26/67 (38%)
Frame = +2
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMI 100
P +P P + PP P SPP SP PP PPP E+
Sbjct: 65 PPPSPPPPHSPPPPPPVYPYLSPPPPPPVHSPPPPVYSPPPPPPSPPPCIES-------- 220
Query: 101 GPPPPPP 107
PPPPPP
Sbjct: 221 -PPPPPP 238
Score = 32.0 bits (71), Expect = 0.38
Identities = 20/56 (35%), Positives = 22/56 (38%), Gaps = 5/56 (8%)
Frame = +2
Query: 57 CLHSIRPPKNPNP-----PSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
C+ S PP P P P PP SP P PPP + PPPP P
Sbjct: 209 CIESPPPPPPPPPCEEHSPPPP----SPHSAPYHPPPSPSPPPPPIQYNSPPPPSP 364
Score = 28.5 bits (62), Expect = 4.2
Identities = 11/24 (45%), Positives = 12/24 (49%)
Frame = +2
Query: 83 PPPPPPHHEADDDDGEMIGPPPPP 106
PPPPPP + PPPPP
Sbjct: 2 PPPPPPVYSPPAPPPTYSPPPPPP 73
>TC219999 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (5%)
Length = 641
Score = 43.5 bits (101), Expect = 1e-04
Identities = 33/89 (37%), Positives = 36/89 (40%)
Frame = -3
Query: 21 KQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPL 80
K S QTP + T SVP P T PS + S+RPP PSP SP
Sbjct: 390 KHSHIQTP--PLSTQTAASVPIPEPST-----PSVQTFLSLRPPT----PSPSPTTSSPS 244
Query: 81 VGPPPPPPHHEADDDDGEMIGPPPPPPGS 109
PPPPPP P PPP S
Sbjct: 243 YPPPPPPPPPH-----------PSPPPSS 190
>TC227037 homologue to UP|MSI4_ARATH (O22607) WD-40 repeat protein MSI4,
partial (86%)
Length = 1945
Score = 43.5 bits (101), Expect = 1e-04
Identities = 26/92 (28%), Positives = 42/92 (45%), Gaps = 2/92 (2%)
Frame = +2
Query: 185 QIEPSEGHQIRNLSWSPTADRFLCVTGSAQ--AKIYDRDGLTLGEFVKGDMYIRDLKNTK 242
++E + + + W+P D L +TGSA +++DR LT + +
Sbjct: 920 KVEKAHNADLHCVDWNPHDDN-LILTGSADNSVRMFDRRNLTTNGVGS------PIHKFE 1078
Query: 243 GHITGLTCGEWHPKTKETILTSSEDGSLRIWD 274
GH + C +W P +S+EDG L IWD
Sbjct: 1079 GHKAAVLCVQWSPDKSSVFGSSAEDGLLNIWD 1174
Score = 37.7 bits (86), Expect = 0.007
Identities = 26/92 (28%), Positives = 44/92 (47%), Gaps = 2/92 (2%)
Frame = +2
Query: 191 GHQ--IRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGL 248
GH+ + ++++ P++ + C G D L L + G + +K K H L
Sbjct: 797 GHEDTVEDVTFCPSSAQEFCSVG-------DDSCLILWDARVGSSPV--VKVEKAHNADL 949
Query: 249 TCGEWHPKTKETILTSSEDGSLRIWDVNDFTT 280
C +W+P ILT S D S+R++D + TT
Sbjct: 950 HCVDWNPHDDNLILTGSADNSVRMFDRRNLTT 1045
>TC209348 similar to UP|Q9LSD7 (Q9LSD7) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MOJ10, partial (12%)
Length = 1405
Score = 43.1 bits (100), Expect = 2e-04
Identities = 21/53 (39%), Positives = 25/53 (46%), Gaps = 4/53 (7%)
Frame = +2
Query: 40 VPTSNPKTTSDDFPSKECLHSIRPPKN----PNPPSPPLQDDSPLVGPPPPPP 88
VP+ NP+ FP E + P N PN P PP P + PPPPPP
Sbjct: 515 VPSQNPQQPLPPFPLSESIQHNPPTTNQQSTPNEPPPPPSSSPPPLPPPPPPP 673
Score = 33.9 bits (76), Expect = 0.099
Identities = 21/64 (32%), Positives = 26/64 (39%), Gaps = 12/64 (18%)
Frame = +2
Query: 60 SIRPPKNPNPPSPP------LQDDSPLV------GPPPPPPHHEADDDDGEMIGPPPPPP 107
S P +NP P PP +Q + P PPPPP + PPPPPP
Sbjct: 509 SFVPSQNPQQPLPPFPLSESIQHNPPTTNQQSTPNEPPPPP-----SSSPPPLPPPPPPP 673
Query: 108 GSNF 111
S +
Sbjct: 674 MSQY 685
>TC232055 similar to UP|O49524 (O49524) Pherophorin - like protein, partial
(7%)
Length = 448
Score = 43.1 bits (100), Expect = 2e-04
Identities = 22/60 (36%), Positives = 25/60 (41%), Gaps = 14/60 (23%)
Frame = +2
Query: 62 RPPKNPNPPSPPLQDDSP--------------LVGPPPPPPHHEADDDDGEMIGPPPPPP 107
R P+ PNPP P SP + PPPPPP A + PPPPPP
Sbjct: 137 RAPRVPNPPPKPTSSSSPSSPSSVSSGETERRIPPPPPPPPRKPASNAAAAPPPPPPPPP 316
Score = 37.0 bits (84), Expect = 0.012
Identities = 22/59 (37%), Positives = 25/59 (42%), Gaps = 6/59 (10%)
Frame = +2
Query: 40 VPTSNPKTTSDDFPSKECLHSIR------PPKNPNPPSPPLQDDSPLVGPPPPPPHHEA 92
VP PK TS PS S PP P PP P + + PPPPPP +A
Sbjct: 149 VPNPPPKPTSSSSPSSPSSVSSGETERRIPPPPPPPPRKPASNAAAAPPPPPPPPPAKA 325
>AI736821 weakly similar to GP|11036868|gb| PxORF73 peptide {Plutella
xylostella granulovirus}, partial (29%)
Length = 430
Score = 43.1 bits (100), Expect = 2e-04
Identities = 28/73 (38%), Positives = 28/73 (38%), Gaps = 6/73 (8%)
Frame = +2
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSP------LVGPPPPPPHHEADD 94
P S P T S F EC S P P PP P SP PPPPPP
Sbjct: 188 PHSPPLTLSARF-GPECPISPLPSPTPPPPITPPSPTSPPPPARERTAPPPPPPPPPPTR 364
Query: 95 DDGEMIGPPPPPP 107
PPPPPP
Sbjct: 365 TTAPRSSPPPPPP 403
Score = 33.1 bits (74), Expect = 0.17
Identities = 22/73 (30%), Positives = 34/73 (46%)
Frame = +2
Query: 16 PLSFGKQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQ 75
PL+ + + P+ + T P + P TS P++E + PP P PP+ +
Sbjct: 200 PLTLSARFGPECPISPLPSPTPPP-PITPPSPTSPPPPARERT-APPPPPPPPPPT---R 364
Query: 76 DDSPLVGPPPPPP 88
+P PPPPPP
Sbjct: 365 TTAPRSSPPPPPP 403
>TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%)
Length = 1048
Score = 42.7 bits (99), Expect = 2e-04
Identities = 29/120 (24%), Positives = 55/120 (45%)
Frame = +2
Query: 156 GSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTADRFLCVTGSAQA 215
G +LS S D +R++ + +N+ L ++ + + ++ +SP F + A
Sbjct: 23 GDFILSSSADSTIRLWSTK-LNANLVCYK----GHNYPVWDVQFSPVGHYFASSSHDRTA 187
Query: 216 KIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSSEDGSLRIWDV 275
+I+ D I+ L+ GH++ + C +WH I T S D ++R+WDV
Sbjct: 188 RIWSMDR------------IQPLRIMAGHLSDVDCVQWHANC-NYIATGSSDKTVRLWDV 328
Score = 32.0 bits (71), Expect = 0.38
Identities = 21/63 (33%), Positives = 32/63 (50%)
Frame = +2
Query: 140 LKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSW 199
L GHT V +LA GS + SGS D V+++D S S + + +++R+L
Sbjct: 479 LIGHTSCVWSLAFSSEGSVIASGSADCTVKLWDVN--TSTKVSRAEEKGGSANRLRSLKT 652
Query: 200 SPT 202
PT
Sbjct: 653 LPT 661
Score = 28.1 bits (61), Expect = 5.4
Identities = 21/79 (26%), Positives = 36/79 (44%)
Frame = +2
Query: 139 VLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLS 198
V GH ++ +LA+ G + SG D + M+D L S R + P GH S
Sbjct: 350 VFVGHRGMILSLAMSPDGRYMASGDEDGTIMMWD-------LSSGRCLTPLIGHTSCVWS 508
Query: 199 WSPTADRFLCVTGSAQAKI 217
+ +++ + +GSA +
Sbjct: 509 LAFSSEGSVIASGSADCTV 565
Score = 27.3 bits (59), Expect = 9.3
Identities = 34/142 (23%), Positives = 56/142 (38%), Gaps = 1/142 (0%)
Frame = +2
Query: 139 VLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLS 198
++ GH V + + + +GS D VR++D +QS + GH+ LS
Sbjct: 224 IMAGHLSDVDCVQWHANCNYIATGSSDKTVRLWD-------VQSGECVRVFVGHRGMILS 382
Query: 199 WSPTADRFLCVTGSAQAKIYDRDG-LTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKT 257
+ + D +G D DG + + + G R L GH T
Sbjct: 383 LAMSPDGRYMASG-------DEDGTIMMWDLSSG----RCLTPLIGH-TSCVWSLAFSSE 526
Query: 258 KETILTSSEDGSLRIWDVNDFT 279
I + S D ++++WDVN T
Sbjct: 527 GSVIASGSADCTVKLWDVNTST 592
>BQ081339 similar to GP|27817931|db P0453E05.6 {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 420
Score = 42.7 bits (99), Expect = 2e-04
Identities = 18/45 (40%), Positives = 23/45 (51%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P + PP PPL + PPPPPP +A ++ PP PPP
Sbjct: 98 PKPSSAPPPPPLLPSNANPPPPPPPPLPKASPSGDSLLQPPTPPP 232
Score = 28.5 bits (62), Expect = 4.2
Identities = 16/45 (35%), Positives = 19/45 (41%), Gaps = 1/45 (2%)
Frame = +2
Query: 45 PKTTSDDFPSKECLHSIRPPKNPNPPSPPLQ-DDSPLVGPPPPPP 88
PK +S P + PP P PP P L+ PP PPP
Sbjct: 98 PKPSSAPPPPPLLPSNANPPPPPPPPLPKASPSGDSLLQPPTPPP 232
Score = 28.5 bits (62), Expect = 4.2
Identities = 18/47 (38%), Positives = 21/47 (44%), Gaps = 2/47 (4%)
Frame = +3
Query: 36 TRRSVP--TSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPL 80
T S+P NP TT P+ L + PP PNPP P S L
Sbjct: 6 TSLSLPFNNENPSTTKTQNPT--ILMRLLPPLRPNPPQPHRHHPSSL 140
>TC214914 homologue to UP|Q8LK71 (Q8LK71) Extensin-like protein, complete
Length = 852
Score = 42.7 bits (99), Expect = 2e-04
Identities = 24/72 (33%), Positives = 30/72 (41%), Gaps = 3/72 (4%)
Frame = -3
Query: 40 VPTSNPKTTSDDFPSKECLHS---IRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDD 96
V +NP +T PS + PP+ P PP PP + P PP PP +
Sbjct: 463 VTFNNPFSTLAHTPSLSASRGQEVLEPPEPPEPPEPPEPPEPPEPPEPPEPPEPPEPPEP 284
Query: 97 GEMIGPPPPPPG 108
E PP PP G
Sbjct: 283 PEPPEPPEPPEG 248
>TC222801 weakly similar to UP|O23370 (O23370) Cell wall protein like,
partial (18%)
Length = 562
Score = 42.7 bits (99), Expect = 2e-04
Identities = 19/45 (42%), Positives = 22/45 (48%)
Frame = +3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP N + P PP P++ PPPPPP PPPPPP
Sbjct: 30 PPTNSSSPPPP---PPPVLSPPPPPPVLSPPPPTNSSSPPPPPPP 155
Score = 38.1 bits (87), Expect = 0.005
Identities = 20/46 (43%), Positives = 24/46 (51%)
Frame = +3
Query: 62 RPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
+PP +PP PP SP PPPPPP ++ PPPPPP
Sbjct: 3 QPPPILSPP-PPTNSSSP---PPPPPP----------VLSPPPPPP 98
Score = 35.0 bits (79), Expect = 0.045
Identities = 21/64 (32%), Positives = 26/64 (39%)
Frame = +3
Query: 43 SNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGP 102
S P T+ P + PP P SPP +S PPPPP ++ P
Sbjct: 21 SPPPPTNSSSPPPPPPPVLSPPPPPPVLSPPPPTNSSSPPPPPPP-----------VLSP 167
Query: 103 PPPP 106
PPPP
Sbjct: 168 PPPP 179
Score = 31.2 bits (69), Expect = 0.64
Identities = 17/52 (32%), Positives = 21/52 (39%)
Frame = +3
Query: 36 TRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPP 87
T S P P P L P + +PP PP P++ PPPPP
Sbjct: 36 TNSSSPPPPPPPVLSPPPPPPVLSPPPPTNSSSPPPPP----PPVLSPPPPP 179
>TC207267 similar to UP|Q8GTM1 (Q8GTM1) WD40, partial (53%)
Length = 1254
Score = 42.4 bits (98), Expect = 3e-04
Identities = 40/136 (29%), Positives = 58/136 (42%), Gaps = 1/136 (0%)
Frame = +2
Query: 142 GHTKVVSALAVDHTGSRVLSG-SYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWS 200
GHT V L T S V + S D + ++D + S SF+ + + +SW+
Sbjct: 347 GHTASVEDLQWSPTESHVFASCSVDGNIAIWDTRLGKSPAASFK----AHNADVNVMSWN 514
Query: 201 PTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKET 260
A L +GS I RD L E GD + + K IT + EW P +
Sbjct: 515 RLASCMLA-SGSDDGTISIRDLRLLKE---GDSVVAHFEYHKHPITSI---EWSPHEASS 673
Query: 261 ILTSSEDGSLRIWDVN 276
+ SS D L IWD++
Sbjct: 674 LAVSSSDNQLTIWDLS 721
>BM954311 weakly similar to GP|21322711|e pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (9%)
Length = 406
Score = 42.4 bits (98), Expect = 3e-04
Identities = 24/78 (30%), Positives = 28/78 (35%), Gaps = 4/78 (5%)
Frame = +2
Query: 33 HKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPN----PPSPPLQDDSPLVGPPPPPP 88
H + ++ P FP H PP P+ PP PP PPPPPP
Sbjct: 20 HNKNNTQMASTQPNNYFPTFPPPSPSHPFNPPPPPSQSFYPPPPPSH-----FNPPPPPP 184
Query: 89 HHEADDDDGEMIGPPPPP 106
H PPPPP
Sbjct: 185 H---------SFSPPPPP 211
Score = 42.4 bits (98), Expect = 3e-04
Identities = 24/56 (42%), Positives = 27/56 (47%), Gaps = 7/56 (12%)
Frame = +2
Query: 59 HSIRPPKNP---NPPSPP--LQDDSPLVGPPPPPP--HHEADDDDGEMIGPPPPPP 107
HS PP P +PPSPP P V PPPPPP H + + PPP PP
Sbjct: 185 HSFSPPPPPFRTSPPSPPRPFSPPPPKVRPPPPPPRPIHPSPPPPPRPVRPPPLPP 352
Score = 38.5 bits (88), Expect = 0.004
Identities = 22/51 (43%), Positives = 23/51 (44%), Gaps = 4/51 (7%)
Frame = +2
Query: 63 PPKNP----NPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGS 109
PP +P NPP PP Q PPPPP H PPPPPP S
Sbjct: 80 PPPSPSHPFNPPPPPSQS----FYPPPPPSH----------FNPPPPPPHS 190
Score = 30.4 bits (67), Expect = 1.1
Identities = 19/49 (38%), Positives = 21/49 (42%)
Frame = +2
Query: 64 PKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFN 112
P N P PP P PPPPP + PPPPP S+FN
Sbjct: 56 PNNYFPTFPPPSPSHPF-NPPPPP---------SQSFYPPPPP--SHFN 166
Score = 30.4 bits (67), Expect = 1.1
Identities = 25/79 (31%), Positives = 30/79 (37%), Gaps = 6/79 (7%)
Frame = +2
Query: 38 RSVPTSNPKTTSDDFPSKECLHSIRPPK------NPNPPSPPLQDDSPLVGPPPPPPHHE 91
R+ P S P+ S P +RPP +P+PP PP V PPP PP
Sbjct: 215 RTSPPSPPRPFSPPPPK------VRPPPPPPRPIHPSPPPPPRP-----VRPPPLPP--- 352
Query: 92 ADDDDGEMIGPPPPPPGSN 110
P PP P N
Sbjct: 353 ---------APLPPSPSPN 382
>TC209891 weakly similar to UP|Q9LW52 (Q9LW52) Arabidopsis thaliana genomic
DNA, chromosome 3, P1 clone: MLM24, partial (10%)
Length = 646
Score = 42.4 bits (98), Expect = 3e-04
Identities = 23/52 (44%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Frame = -3
Query: 58 LHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPP-PPPG 108
L + RP +NP PP PP + P PP PPP G+ PPP PPPG
Sbjct: 335 LKNPRPGRNPPPPMPPPGKNPPPPPPPMPPP--------GKYPAPPPMPPPG 204
Score = 35.4 bits (80), Expect = 0.034
Identities = 15/41 (36%), Positives = 18/41 (43%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPP 103
P KNP PP PP+ PPP PP + + PP
Sbjct: 287 PGKNPPPPPPPMPPPGKYPAPPPMPPPGKKLPPPNSLASPP 165
>BQ630253
Length = 429
Score = 42.0 bits (97), Expect = 4e-04
Identities = 26/70 (37%), Positives = 30/70 (42%), Gaps = 6/70 (8%)
Frame = +1
Query: 44 NPKTTSDDFPSKECLHSIRPPKNPNPP------SPPLQDDSPLVGPPPPPPHHEADDDDG 97
+P S P HS PP +P+PP SPP SP P PPPP+H
Sbjct: 166 SPPPPSPSPPPPYYYHS-PPPPSPSPPPPYYYKSPPPPSPSPPPSPSPPPPYH------- 321
Query: 98 EMIGPPPPPP 107
PPPP P
Sbjct: 322 -YTSPPPPSP 348
Score = 39.7 bits (91), Expect = 0.002
Identities = 23/67 (34%), Positives = 28/67 (41%), Gaps = 3/67 (4%)
Frame = +1
Query: 44 NPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPP 103
+P S P HS PP +P+PP P P P PPPP++ PP
Sbjct: 118 SPPPPSPSPPPPYYYHS-PPPPSPSPPPPYYYHSPPPPSPSPPPPYYYKSPPPPSPSPPP 294
Query: 104 ---PPPP 107
PPPP
Sbjct: 295 SPSPPPP 315
Score = 38.9 bits (89), Expect = 0.003
Identities = 23/67 (34%), Positives = 28/67 (41%), Gaps = 10/67 (14%)
Frame = +1
Query: 53 PSKECLHS-----IRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPP--- 104
P KE +H PP +P+PP P P P PPPP++ PPP
Sbjct: 31 PPKEHVHPPYYYHSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYHSPPPPSPSPPPPYYY 210
Query: 105 --PPPGS 109
PPP S
Sbjct: 211 HSPPPPS 231
Score = 30.8 bits (68), Expect = 0.84
Identities = 15/45 (33%), Positives = 18/45 (39%)
Frame = +1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +P+PP P P P P P + I PPPP
Sbjct: 286 PPPSPSPPPPYHYTSPPPPSPAPAPKYIYKSPPPPVYIYASPPPP 420
Score = 27.7 bits (60), Expect = 7.1
Identities = 17/47 (36%), Positives = 18/47 (38%)
Frame = +1
Query: 59 HSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPP 105
HS PPK PP SP P PPP + PPPP
Sbjct: 19 HSPPPPKEH--VHPPYYYHSPPPPSPSPPPPYYYKSPPPPSPSPPPP 153
>TC224723 hydroxyproline-rich glycoprotein
Length = 2345
Score = 42.0 bits (97), Expect = 4e-04
Identities = 24/67 (35%), Positives = 30/67 (43%)
Frame = +1
Query: 44 NPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPP 103
+P S P HS PP +P+PPSP P P PPPP++ PP
Sbjct: 928 SPPPPSPSPPPPYYYHS-PPPPSPSPPSPYYYKSPPPPSPSPPPPYYYQ--------SPP 1080
Query: 104 PPPPGSN 110
PP P S+
Sbjct: 1081PPSPTSH 1101
Score = 41.2 bits (95), Expect = 6e-04
Identities = 21/56 (37%), Positives = 25/56 (44%), Gaps = 5/56 (8%)
Frame = +1
Query: 59 HSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPP-----PPPGS 109
HS PP +P+PP P P P PPPP++ PPP PPP S
Sbjct: 826 HSPPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYHSPPPPSPSPPPPYYYHSPPPPS 993
Score = 35.8 bits (81), Expect = 0.026
Identities = 23/71 (32%), Positives = 28/71 (39%), Gaps = 5/71 (7%)
Frame = +1
Query: 44 NPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPP 103
+P S P HS PP +P+PP P P P PP P++ PP
Sbjct: 880 SPPPPSPSPPPPYYYHSPPPP-SPSPPPPYYYHSPPPPSPSPPSPYYYKSPPPPSPSPPP 1056
Query: 104 P-----PPPGS 109
P PPP S
Sbjct: 1057PYYYQSPPPPS 1089
Score = 34.7 bits (78), Expect = 0.058
Identities = 17/45 (37%), Positives = 18/45 (39%)
Frame = +1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP PP P P P PPPP+H PPPP P
Sbjct: 1126 PPPTSYPPPPYHYVSPPPPSPSPPPPYHYTS--------PPPPSP 1236
Score = 34.7 bits (78), Expect = 0.058
Identities = 17/50 (34%), Positives = 21/50 (42%), Gaps = 5/50 (10%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPP-----PPPP 107
PP +P+PP P P P P PP++ PP PPPP
Sbjct: 546 PPPSPSPPPPYYYQSPPPPSPTPHPPYYYKSPPPPTSHPPPYHYVSPPPP 397
Score = 33.5 bits (75), Expect = 0.13
Identities = 25/81 (30%), Positives = 32/81 (38%), Gaps = 1/81 (1%)
Frame = +1
Query: 28 PLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPP-PP 86
P E +H P P + P + PP +P+PP PP SP P PP
Sbjct: 793 PKEHVHPPYYYHSPPPPPSPS----PPPPYYYKSPPPPSPSPP-PPYYYHSPPPPSPSPP 957
Query: 87 PPHHEADDDDGEMIGPPPPPP 107
PP++ PPPP P
Sbjct: 958 PPYY--------YHSPPPPSP 996
Score = 33.1 bits (74), Expect = 0.17
Identities = 20/48 (41%), Positives = 23/48 (47%), Gaps = 3/48 (6%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPP---PPPHHEADDDDGEMIGPPPPPP 107
PP +P P PP SP PPP PPP+H + PPPP P
Sbjct: 498 PPPSP-TPHPPYYYKSP---PPPTSHPPPYH--------YVSPPPPSP 391
Score = 33.1 bits (74), Expect = 0.17
Identities = 22/63 (34%), Positives = 26/63 (40%), Gaps = 18/63 (28%)
Frame = +1
Query: 63 PPKNPNPP------SPPLQDDSPLVGPP------------PPPPHHEADDDDGEMIGPPP 104
PP +P+PP SPP SP PP PPPP+H + PPP
Sbjct: 1030 PPPSPSPPPPYYYQSPP--PPSPTSHPPYYYKSPPPPTSYPPPPYH--------YVSPPP 1179
Query: 105 PPP 107
P P
Sbjct: 1180 PSP 1188
Score = 33.1 bits (74), Expect = 0.17
Identities = 22/71 (30%), Positives = 25/71 (34%), Gaps = 5/71 (7%)
Frame = +1
Query: 44 NPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPP 103
+P S P HS PPK P P P PPPP++ PP
Sbjct: 733 SPPPPSPSPPPPYYYHSPPPPKEHVHPPYYYHSPPPPPSPSPPPPYYYKSPPPPSPSPPP 912
Query: 104 P-----PPPGS 109
P PPP S
Sbjct: 913 PYYYHSPPPPS 945
Score = 31.2 bits (69), Expect = 0.64
Identities = 20/65 (30%), Positives = 25/65 (37%)
Frame = +1
Query: 43 SNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGP 102
S P TS +P + PP +P+PP P P P P P + I
Sbjct: 1120 SPPPPTS--YPPPPYHYVSPPPPSPSPPPPYHYTSPPPPSPAPAPKYIYKSPPPPVYIYA 1293
Query: 103 PPPPP 107
PPPP
Sbjct: 1294 SPPPP 1308
Score = 30.8 bits (68), Expect = 0.84
Identities = 19/47 (40%), Positives = 22/47 (46%), Gaps = 4/47 (8%)
Frame = -3
Query: 65 KNPNPPS---PPLQDDSPLVGPP-PPPPHHEADDDDGEMIGPPPPPP 107
K+P PP+ PP SP P PPPP+H PPPP P
Sbjct: 459 KSPPPPTSHPPPYHYVSPPPPSPSPPPPYH--------YTSPPPPSP 343
Score = 30.8 bits (68), Expect = 0.84
Identities = 15/45 (33%), Positives = 18/45 (39%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +P+PP P P P P P + I PPPP
Sbjct: 405 PPPSPSPPPPYHYTSPPPPSPAPAPKYIYKSPPPPVYIYASPPPP 271
Score = 30.0 bits (66), Expect = 1.4
Identities = 14/40 (35%), Positives = 17/40 (42%)
Frame = -3
Query: 68 NPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
+PP P P P PPPP++ PPPP P
Sbjct: 579 SPPPPYYYKSPPPPSPSPPPPYYYQ--------SPPPPSP 484
Score = 28.1 bits (61), Expect = 5.4
Identities = 20/66 (30%), Positives = 25/66 (37%), Gaps = 2/66 (3%)
Frame = -3
Query: 43 SNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPH--HEADDDDGEMI 100
S P TS P H + PP P PP SP P P P +++ +
Sbjct: 456 SPPPPTSHPPP----YHYVSPPPPSPSPPPPYHYTSPPPPSPAPAPKYIYKSPPPPVYIY 289
Query: 101 GPPPPP 106
PPPP
Sbjct: 288 ASPPPP 271
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,080,060
Number of Sequences: 63676
Number of extensions: 460483
Number of successful extensions: 16006
Number of sequences better than 10.0: 1123
Number of HSP's better than 10.0 without gapping: 5381
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9605
length of query: 316
length of database: 12,639,632
effective HSP length: 97
effective length of query: 219
effective length of database: 6,463,060
effective search space: 1415410140
effective search space used: 1415410140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146819.11


