

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146819.1 + phase: 0
(221 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
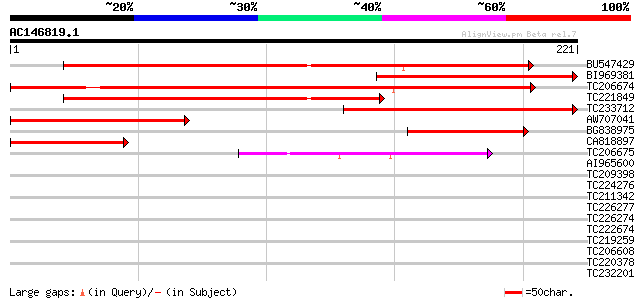
Score E
Sequences producing significant alignments: (bits) Value
BU547429 213 7e-56
BI969381 160 3e-40
TC206674 similar to UP|FMO3_HUMAN (P31513) Dimethylaniline monoo... 157 3e-39
TC221849 143 5e-35
TC233712 similar to UP|O23024 (O23024) T1G11.14 protein (Flavin-... 135 2e-32
AW707041 121 2e-28
BG838975 similar to GP|16555354|gb| flavin-containing monooxygen... 68 4e-12
CA818897 similar to GP|16555354|gb flavin-containing monooxygena... 62 3e-10
TC206675 56 1e-08
AI965600 40 0.001
TC209398 similar to GB|AAP13398.1|30023730|BT006290 At1g25280 {A... 30 0.86
TC224276 28 2.5
TC211342 similar to UP|Q9LVF2 (Q9LVF2) Arabidopsis thaliana geno... 28 2.5
TC226277 weakly similar to UP|Q6K2F0 (Q6K2F0) Heat shock protein... 28 3.3
TC226274 similar to UP|Q8L5W5 (Q8L5W5) VERNALIZATION INDEPENDENC... 28 3.3
TC222674 similar to UP|Q6PSU8 (Q6PSU8) Formin homology 2 domain-... 27 5.5
TC219259 similar to UP|Q9S7V4 (Q9S7V4) AtHVA22a; 65476-64429 (At... 27 7.2
TC206608 UP|Q8LJR3 (Q8LJR3) 14-.3.3 protein, complete 27 7.2
TC220378 27 7.2
TC232201 27 9.5
>BU547429
Length = 665
Score = 213 bits (541), Expect = 7e-56
Identities = 101/184 (54%), Positives = 140/184 (75%), Gaps = 1/184 (0%)
Frame = -2
Query: 22 VHVLPREIFGISTFELAVMMLKWLPLWIVDKLLLILTWFILGDMEKYGIKRPSMGPLQLK 81
VH+LP+++ G STF L++ +LKW P+ VD+ LL+++ + D ++G++RP +GPL+LK
Sbjct: 580 VHILPQQMXGKSTFGLSMFLLKWFPIRFVDQFLLLMSHLMXXDTAQFGLRRPKLGPLELK 401
Query: 82 NTVGKTPVLDIGALEKIRSGDINVVPGIKRINKNGEVELVNGEKLDIDAVVLATGYRSNV 141
N GKTPVLD+G L +I++G I V GIKR+ +N VE V+G+ + DA+VLATGY+SNV
Sbjct: 400 NLYGKTPVLDVGTLTQIKNGKIKVCRGIKRLARNA-VEFVDGKVENFDAMVLATGYKSNV 224
Query: 142 PSWLQEGEFFS-KNGYPKMPFPHGWKGNSGLYAVGFTKRGLSGASSDAVKIAQDIGKVWK 200
PSWL+ + FS K+G+P+ PFP+GWKG +GLYAVGFTKRGL GAS DA +IA+DI WK
Sbjct: 223 PSWLKGSDMFSEKDGFPRKPFPNGWKGENGLYAVGFTKRGLLGASIDAKRIAEDIEHSWK 44
Query: 201 QETK 204
E K
Sbjct: 43 AEAK 32
>BI969381
Length = 602
Score = 160 bits (406), Expect = 3e-40
Identities = 73/78 (93%), Positives = 77/78 (98%)
Frame = -2
Query: 144 WLQEGEFFSKNGYPKMPFPHGWKGNSGLYAVGFTKRGLSGASSDAVKIAQDIGKVWKQET 203
++QEGEFFSKNGYPKMPFPHGWKGN+GLYAVGFTKRGLSGASSDAVKIAQDIG+VWK ET
Sbjct: 544 YVQEGEFFSKNGYPKMPFPHGWKGNAGLYAVGFTKRGLSGASSDAVKIAQDIGQVWKNET 365
Query: 204 KQKKQRTTACHRRCISQF 221
KQKKQRTTACHRRCISQF
Sbjct: 364 KQKKQRTTACHRRCISQF 311
>TC206674 similar to UP|FMO3_HUMAN (P31513) Dimethylaniline monooxygenase
[N-oxide-forming] 3 (Hepatic flavin-containing
monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3)
(FMO form 2) (FMO II) , partial (4%)
Length = 1470
Score = 157 bits (398), Expect = 3e-39
Identities = 79/206 (38%), Positives = 129/206 (62%), Gaps = 1/206 (0%)
Frame = +3
Query: 1 MELSLDLSNHHALPSMVVRSSVHVLPREIFGISTFELAVMMLKWLPLWIVDKLLLILTWF 60
ME++LDLSN A PS++VRS VH L R++ A +ML +L L V+K+L++++
Sbjct: 588 MEIALDLSNFGAKPSIIVRSPVHFLSRDMM-----YYASLMLNYLSLSTVEKVLVMVSKV 752
Query: 61 ILGDMEKYGIKRPSMGPLQLKNTVGKTPVLDIGALEKIRSGDINVVPGIKRINKNGEVEL 120
+ GD+ +YGI PS GP +K K P++D+G ++KI+S +I V+P + + EV
Sbjct: 753 VYGDLSEYGIPYPSEGPFTMKMKYAKFPIIDVGTVKKIKSREIQVLPAEIKSIRGNEVLF 932
Query: 121 VNGEKLDIDAVVLATGYRSNVPSWLQEG-EFFSKNGYPKMPFPHGWKGNSGLYAVGFTKR 179
+G+ D++V TG++ + WL+ G + +++G+PK FP+ WKG +GLY VG ++R
Sbjct: 933 QDGKSYTFDSIVFCTGFKRSTQKWLKGGDDLLNEDGFPKNSFPNHWKGRNGLYCVGLSRR 1112
Query: 180 GLSGASSDAVKIAQDIGKVWKQETKQ 205
G GA+ DA +A DI + QE ++
Sbjct: 1113GFFGANMDAQLVANDIASLIPQEERE 1190
>TC221849
Length = 452
Score = 143 bits (361), Expect = 5e-35
Identities = 66/125 (52%), Positives = 97/125 (76%)
Frame = +1
Query: 22 VHVLPREIFGISTFELAVMMLKWLPLWIVDKLLLILTWFILGDMEKYGIKRPSMGPLQLK 81
VH+LP+++ G STF L++ +LKW P+ VD+ LL+++ +LGD ++G++RP +GPL+LK
Sbjct: 73 VHILPQQMLGKSTFGLSMFLLKWFPIRFVDQFLLLMSHLMLGDTAQFGLRRPKLGPLELK 252
Query: 82 NTVGKTPVLDIGALEKIRSGDINVVPGIKRINKNGEVELVNGEKLDIDAVVLATGYRSNV 141
N GKTPVLD+G L +I++G I V GIKR+ +N VE V+G+ + DA+VLATGY+SNV
Sbjct: 253 NLYGKTPVLDVGTLTQIKNGKIKVCRGIKRLARNA-VEFVDGKVENFDAMVLATGYKSNV 429
Query: 142 PSWLQ 146
PSWL+
Sbjct: 430 PSWLK 444
>TC233712 similar to UP|O23024 (O23024) T1G11.14 protein (Flavin-containing
monooxygenase YUCCA3) (At1g04610), partial (21%)
Length = 741
Score = 135 bits (339), Expect = 2e-32
Identities = 59/91 (64%), Positives = 73/91 (79%)
Frame = +3
Query: 131 VVLATGYRSNVPSWLQEGEFFSKNGYPKMPFPHGWKGNSGLYAVGFTKRGLSGASSDAVK 190
VVLATGY SNVPSWL+E +FF+ +G P+ PFP+GW+G GLYAVGFT++GLSGAS DA+
Sbjct: 18 VVLATGYHSNVPSWLKENDFFTSDGTPRNPFPNGWRGKGGLYAVGFTRKGLSGASLDAIN 197
Query: 191 IAQDIGKVWKQETKQKKQRTTACHRRCISQF 221
+A DI K WK+E KQK++ A HRRCIS F
Sbjct: 198 VAHDIAKNWKEENKQKRKTVAARHRRCISHF 290
>AW707041
Length = 365
Score = 121 bits (304), Expect = 2e-28
Identities = 55/70 (78%), Positives = 65/70 (92%)
Frame = +1
Query: 1 MELSLDLSNHHALPSMVVRSSVHVLPREIFGISTFELAVMMLKWLPLWIVDKLLLILTWF 60
MELSLDL NHH+ PSMVVRSSVHVLPRE+FGIS FELAVM+L+WLPLW+VDK+LLIL WF
Sbjct: 154 MELSLDLCNHHSSPSMVVRSSVHVLPREVFGISIFELAVMLLQWLPLWLVDKILLILAWF 333
Query: 61 ILGDMEKYGI 70
+LG++EK G+
Sbjct: 334 VLGNIEKLGL 363
>BG838975 similar to GP|16555354|gb| flavin-containing monooxygenase YUCCA2
{Arabidopsis thaliana}, partial (13%)
Length = 428
Score = 67.8 bits (164), Expect = 4e-12
Identities = 29/47 (61%), Positives = 35/47 (73%)
Frame = -2
Query: 156 YPKMPFPHGWKGNSGLYAVGFTKRGLSGASSDAVKIAQDIGKVWKQE 202
+P+ PFP+GWKG +G Y VGFTK GL GAS DA +IA+DI WK E
Sbjct: 319 FPRKPFPNGWKGENGXYXVGFTKXGLLGASIDAKRIAEDIEHXWKXE 179
>CA818897 similar to GP|16555354|gb flavin-containing monooxygenase YUCCA2
{Arabidopsis thaliana}, partial (26%)
Length = 445
Score = 61.6 bits (148), Expect = 3e-10
Identities = 26/46 (56%), Positives = 37/46 (79%)
Frame = +1
Query: 1 MELSLDLSNHHALPSMVVRSSVHVLPREIFGISTFELAVMMLKWLP 46
ME+ LDL NH+A PS+VVR +VH+LP+++ G STF L++ +LKW P
Sbjct: 307 MEVCLDLCNHNARPSLVVRDTVHILPQQMLGKSTFGLSMFLLKWFP 444
>TC206675
Length = 957
Score = 56.2 bits (134), Expect = 1e-08
Identities = 35/104 (33%), Positives = 56/104 (53%), Gaps = 5/104 (4%)
Frame = +2
Query: 90 LDIGALEKIRSGDINVVPGIKRINKNGEVELVNGEKLD---IDAVVLATGYRSNVPSWLQ 146
+D+G ++KI+S +I V+P + K EV E + + A R + S +
Sbjct: 584 IDVGTVKKIKSREIQVLPA-EIDRKVSEVMKFCSEMANHAHSTQLFSAHASRDQLKSGFR 760
Query: 147 E--GEFFSKNGYPKMPFPHGWKGNSGLYAVGFTKRGLSGASSDA 188
+++NG+PK FP+ WKG +GLY VG ++RG GA+ DA
Sbjct: 761 G**SSKYNENGFPKSSFPNHWKGRNGLYCVGLSRRGFFGANMDA 892
Score = 28.9 bits (63), Expect = 1.9
Identities = 13/25 (52%), Positives = 19/25 (76%)
Frame = +2
Query: 1 MELSLDLSNHHALPSMVVRSSVHVL 25
ME++LDLS+ A PS++VRS + L
Sbjct: 344 MEIALDLSDFGAKPSIIVRSPIGTL 418
>AI965600
Length = 346
Score = 39.7 bits (91), Expect = 0.001
Identities = 16/33 (48%), Positives = 23/33 (69%)
Frame = +2
Query: 61 ILGDMEKYGIKRPSMGPLQLKNTVGKTPVLDIG 93
+L D+ KYG+ RP+ GP +K GK PV+D+G
Sbjct: 245 VLLDVTKYGVARPNEGPFYMKVKYGKYPVIDVG 343
>TC209398 similar to GB|AAP13398.1|30023730|BT006290 At1g25280 {Arabidopsis
thaliana;} , partial (36%)
Length = 650
Score = 30.0 bits (66), Expect = 0.86
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 2/67 (2%)
Frame = +2
Query: 7 LSNHHALPSMVVRSSVHVLPR--EIFGISTFELAVMMLKWLPLWIVDKLLLILTWFILGD 64
L +H +L S+++ S L R I+ + T ++ ++ + LP L+L W + D
Sbjct: 179 LCHHRSLLSLIMTRSFFSLARLARIYLLWTIDIPYLLFRLLPF---A*LVLTQNWLVNSD 349
Query: 65 MEKYGIK 71
E+YG K
Sbjct: 350 NERYGTK 370
>TC224276
Length = 586
Score = 28.5 bits (62), Expect = 2.5
Identities = 21/58 (36%), Positives = 29/58 (49%), Gaps = 3/58 (5%)
Frame = -3
Query: 7 LSNHHALPSMVVRSSVHVLPREIFGISTFELAVMMLKWLPLWIV---DKLLLILTWFI 61
LSNHH + VV + V V+ + G S+ E + +L P WIV DK L F+
Sbjct: 317 LSNHHYVDFHVVDTCVRVVYSLLRGESSLEPNLYILLISPKWIVLNKDKATTQLQMFL 144
>TC211342 similar to UP|Q9LVF2 (Q9LVF2) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MIL23, partial (29%)
Length = 862
Score = 28.5 bits (62), Expect = 2.5
Identities = 11/24 (45%), Positives = 18/24 (74%)
Frame = +2
Query: 39 VMMLKWLPLWIVDKLLLILTWFIL 62
+MML WL L ++ +LL+L+W +L
Sbjct: 221 MMMLLWLQLALMLNILLLLSWIVL 292
>TC226277 weakly similar to UP|Q6K2F0 (Q6K2F0) Heat shock protein-like,
partial (6%)
Length = 730
Score = 28.1 bits (61), Expect = 3.3
Identities = 15/48 (31%), Positives = 25/48 (51%)
Frame = -3
Query: 15 SMVVRSSVHVLPREIFGISTFELAVMMLKWLPLWIVDKLLLILTWFIL 62
S+V+ + VH + G + ++ K LP KLL++L WF+L
Sbjct: 563 SLVLITIVHNRHPPMGGCFPWRRVLIRFKSLPFGTASKLLVLLLWFLL 420
>TC226274 similar to UP|Q8L5W5 (Q8L5W5) VERNALIZATION INDEPENDENCE 4, partial
(4%)
Length = 664
Score = 28.1 bits (61), Expect = 3.3
Identities = 15/48 (31%), Positives = 25/48 (51%)
Frame = +2
Query: 15 SMVVRSSVHVLPREIFGISTFELAVMMLKWLPLWIVDKLLLILTWFIL 62
S+V+ + VH + G + ++ K LP KLL++L WF+L
Sbjct: 356 SLVLITIVHNRHPPMGGCFPWRRVLIRFKSLPFGTASKLLVLLLWFLL 499
>TC222674 similar to UP|Q6PSU8 (Q6PSU8) Formin homology 2 domain-containing
protein 5, partial (15%)
Length = 613
Score = 27.3 bits (59), Expect = 5.5
Identities = 15/41 (36%), Positives = 21/41 (50%)
Frame = +2
Query: 80 LKNTVGKTPVLDIGALEKIRSGDINVVPGIKRINKNGEVEL 120
+K T GKT +L LE IRS I + K K+ ++L
Sbjct: 290 VKGTDGKTTLLHFVVLEIIRSEGIKAIRKAKESQKSSSIKL 412
>TC219259 similar to UP|Q9S7V4 (Q9S7V4) AtHVA22a; 65476-64429 (AtHVA22a),
partial (86%)
Length = 787
Score = 26.9 bits (58), Expect = 7.2
Identities = 12/32 (37%), Positives = 21/32 (65%), Gaps = 1/32 (3%)
Frame = +3
Query: 32 ISTFELA-VMMLKWLPLWIVDKLLLILTWFIL 62
I+ FEL +L+W+P+W KL+L +W ++
Sbjct: 246 ITLFELTFAKVLEWIPIWPYAKLIL-TSWLVI 338
>TC206608 UP|Q8LJR3 (Q8LJR3) 14-.3.3 protein, complete
Length = 1344
Score = 26.9 bits (58), Expect = 7.2
Identities = 20/63 (31%), Positives = 29/63 (45%), Gaps = 1/63 (1%)
Frame = -2
Query: 8 SNHHALPSMVVRSSVHVLPREIFGISTFELAVMMLKWLPLWIVDKLLLILTWFILGDME- 66
S +PS ++ SS +LP F +M W L + LLL ++ GDME
Sbjct: 707 SRRFKIPSQILLSSASILPLYSF---------IMATWSSLPLFSLLLLSSSFSFSGDMES 555
Query: 67 KYG 69
K+G
Sbjct: 554 KWG 546
>TC220378
Length = 715
Score = 26.9 bits (58), Expect = 7.2
Identities = 16/52 (30%), Positives = 27/52 (51%)
Frame = +3
Query: 16 MVVRSSVHVLPREIFGISTFELAVMMLKWLPLWIVDKLLLILTWFILGDMEK 67
+++ S+ L FG + + V++LKWL L+I L+ +L MEK
Sbjct: 534 LIMLPSIIFLYSLAFGFRSLKTVVVILKWL*LYISQL*LIFSGLIVLMIMEK 689
>TC232201
Length = 783
Score = 26.6 bits (57), Expect = 9.5
Identities = 16/36 (44%), Positives = 21/36 (57%), Gaps = 1/36 (2%)
Frame = +3
Query: 2 ELSLDLSNHHALPSMVVRSSVHV-LPREIFGISTFE 36
E D+SNHH S RSS H LP +FG S+++
Sbjct: 66 EEEYDMSNHHPFFSPTSRSSSHASLP--VFGSSSYK 167
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,434,087
Number of Sequences: 63676
Number of extensions: 138811
Number of successful extensions: 751
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 743
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 746
length of query: 221
length of database: 12,639,632
effective HSP length: 94
effective length of query: 127
effective length of database: 6,654,088
effective search space: 845069176
effective search space used: 845069176
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146819.1


