

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146817.8 + phase: 0
(831 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
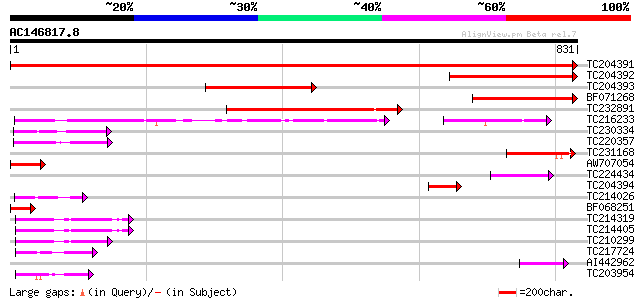
Score E
Sequences producing significant alignments: (bits) Value
TC204391 homologue to UP|Q9ASR1 (Q9ASR1) At1g56070/T6H22_13 (Elo... 1489 0.0
TC204392 homologue to UP|EF2_BETVU (O23755) Elongation factor 2 ... 313 2e-85
TC204393 homologue to UP|Q8RVT3 (Q8RVT3) Elongation factor EF-2 ... 308 8e-84
BF071268 similar to SP|O23755|EF2_ Elongation factor 2 (EF-2). [... 205 6e-53
TC232891 similar to UP|Q9LNC5 (Q9LNC5) F9P14.8 protein, partial ... 197 1e-50
TC216233 GB|CAA50573.1|402753|GMFUSA translation elongation fact... 130 2e-30
TC230334 similar to UP|Q8RWP3 (Q8RWP3) GTP-binding protein LepA-... 101 1e-21
TC220357 homologue to UP|Q6SYB1 (Q6SYB1) GTP-binding protein Typ... 99 1e-20
TC231168 homologue to UP|Q9LNC5 (Q9LNC5) F9P14.8 protein, partia... 92 9e-19
AW707054 similar to GP|15450763|gb| elongation factor EF-2 {Arab... 83 4e-16
TC224434 similar to UP|Q9LS91 (Q9LS91) Elongation factor EF-2, p... 74 2e-13
TC204394 similar to UP|EF2_BETVU (O23755) Elongation factor 2 (E... 71 2e-12
TC214026 similar to UP|Q6G0R7 (Q6G0R7) GTP-binding protein typA,... 64 3e-10
BF068251 similar to GP|15450763|gb| elongation factor EF-2 {Arab... 59 9e-09
TC214319 homologue to UP|EFT1_SOYBN (Q43467) Elongation factor T... 58 2e-08
TC214405 homologue to UP|EFT1_SOYBN (Q43467) Elongation factor T... 58 2e-08
TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor T... 56 6e-08
TC217724 homologue to UP|EFTM_ARATH (Q9ZT91) Elongation factor T... 52 8e-07
AI442962 weakly similar to GP|10798636|emb elongation factor 2 {... 50 4e-06
TC203954 homologue to UP|EF1A_LYCES (P17786) Elongation factor 1... 48 2e-05
>TC204391 homologue to UP|Q9ASR1 (Q9ASR1) At1g56070/T6H22_13 (Elongation
factor EF-2), complete
Length = 2941
Score = 1489 bits (3854), Expect = 0.0
Identities = 728/831 (87%), Positives = 777/831 (92%)
Frame = +1
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD KHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 157 MDYKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 336
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEM+D LK+FKGER GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV
Sbjct: 337 TGISLYYEMTDEALKSFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 516
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL +D EEAY T QRVIE+ NV+MATY
Sbjct: 517 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFQRVIENANVIMATY 696
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LLGDVQVYPEKGTV+FSAGLHGW+FTLTNFAKMYASKFGVDE KMM RLWGENFFD
Sbjct: 697 EDPLLGDVQVYPEKGTVAFSAGLHGWAFTLTNFAKMYASKFGVDEGKMMERLWGENFFDP 876
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+TKKWT+K+T + TCKRGFVQFCYEPIKQII CMNDQKDKLWPMLQKLGV +KSEEK+L
Sbjct: 877 ATKKWTSKNTGSATCKRGFVQFCYEPIKQIINTCMNDQKDKLWPMLQKLGVTMKSEEKDL 1056
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
GKALMKRVMQ+WLPASSALLEMMIFHLPSP+ AQKYRVENLYEGPLDD YASAIRNCDP
Sbjct: 1057MGKALMKRVMQTWLPASSALLEMMIFHLPSPSTAQKYRVENLYEGPLDDQYASAIRNCDP 1236
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
EGPLMLYVSKMIPASDKGRF+AFGRVFSG+VSTG+KVRIMGPNY+PGEKKDLYVKSVQRT
Sbjct: 1237EGPLMLYVSKMIPASDKGRFFAFGRVFSGRVSTGLKVRIMGPNYVPGEKKDLYVKSVQRT 1416
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGK+QETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVV VA
Sbjct: 1417VIWMGKRQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVRVA 1596
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMVVCTI E+GEHI+A AGELHLEICLKDLQDDFM G
Sbjct: 1597VQCKVASDLPKLVEGLKRLAKSDPMVVCTIEESGEHIVAGAGELHLEICLKDLQDDFMGG 1776
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI KSDP+VSFRETVLE+S TVMSKSPNKHNRLYMEARP+EEGLAEAIDDG+IGPRD+
Sbjct: 1777AEIIKSDPVVSFRETVLERSCRTVMSKSPNKHNRLYMEARPLEEGLAEAIDDGKIGPRDD 1956
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PK KILS+EFGWDKDLAKK+WCFGPET GPNM+VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 1957PKVRSKILSEEFGWDKDLAKKIWCFGPETLGPNMVVDMCKGVQYLNEIKDSVVAGFQWAS 2136
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEG +A+EN+R +CFEVCDVVLH DAIHRGGGQIIPTARRVFYA+ +TAKPRLLEPVYLV
Sbjct: 2137KEGALAEENMRAICFEVCDVVLHADAIHRGGGQIIPTARRVFYASQITAKPRLLEPVYLV 2316
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPVIESF F+ +LRA T GQ
Sbjct: 2317EIQAPEQALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVIESFGFSSTLRAATSGQ 2496
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQ VFDHWDM+ SDPLE G+ AA V +IRK+KGLKEQ+ PLSEFED+L
Sbjct: 2497AFPQCVFDHWDMMSSDPLEAGSQAAQLVTDIRKRKGLKEQMTPLSEFEDKL 2649
>TC204392 homologue to UP|EF2_BETVU (O23755) Elongation factor 2 (EF-2),
partial (22%)
Length = 818
Score = 313 bits (802), Expect = 2e-85
Identities = 151/187 (80%), Positives = 168/187 (89%)
Frame = +2
Query: 645 LNEIKDSVVAGFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYA 704
LNEIKDSVVAGFQ ASKEG +A+EN+R +CFEVCDVVLH DAIHRGGGQIIPTARRVFYA
Sbjct: 2 LNEIKDSVVAGFQWASKEGALAEENMRAICFEVCDVVLHADAIHRGGGQIIPTARRVFYA 181
Query: 705 AMLTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVI 764
+ +TAKPRLLEPVYLVEIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPVI
Sbjct: 182 SQITAKPRLLEPVYLVEIQAPEQALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVI 361
Query: 765 ESFQFNESLRAQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPL 824
ESF F+ +LRA T GQAFPQ VFDHWDM+ SDPLE G+ AA V +IRK+KGLKEQ+ PL
Sbjct: 362 ESFGFSSTLRAATSGQAFPQCVFDHWDMMSSDPLEAGSQAAQLVTDIRKRKGLKEQMTPL 541
Query: 825 SEFEDRL 831
SEFED+L
Sbjct: 542 SEFEDKL 562
>TC204393 homologue to UP|Q8RVT3 (Q8RVT3) Elongation factor EF-2 (Fragment),
partial (33%)
Length = 487
Score = 308 bits (788), Expect = 8e-84
Identities = 148/162 (91%), Positives = 157/162 (96%)
Frame = +1
Query: 288 KLGVNLKSEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPL 347
KLGV +KSEEK+L GKALMKRVMQ+WLPASSALLEMMIFHLPSP+ AQKYRVENLYEGPL
Sbjct: 1 KLGVTMKSEEKDLMGKALMKRVMQTWLPASSALLEMMIFHLPSPSTAQKYRVENLYEGPL 180
Query: 348 DDPYASAIRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPG 407
DD YASAIRNCDPEGPLMLYVSKMIPASDKGRF+AFGRVFSG+VSTG+KVRIMGPNY+PG
Sbjct: 181 DDQYASAIRNCDPEGPLMLYVSKMIPASDKGRFFAFGRVFSGRVSTGLKVRIMGPNYVPG 360
Query: 408 EKKDLYVKSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFI 449
EKKDLYVKSVQRTVIWMGK+QET EDVPCGNTVAMVGLDQFI
Sbjct: 361 EKKDLYVKSVQRTVIWMGKRQETAEDVPCGNTVAMVGLDQFI 486
>BF071268 similar to SP|O23755|EF2_ Elongation factor 2 (EF-2). [Sugar beet]
{Beta vulgaris}, partial (18%)
Length = 460
Score = 205 bits (522), Expect = 6e-53
Identities = 103/153 (67%), Positives = 119/153 (77%)
Frame = +2
Query: 679 DVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQ 738
+VVLH DAIH GGG IIPT +R FYA +TA+P LL+PV L EI A E ALGGIY+VLN
Sbjct: 2 NVVLHADAIHTGGGHIIPTVKRPFYANQITAQPILLDPVSLFEIHATELALGGIYTVLNH 181
Query: 739 KRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQAFPQLVFDHWDMVPSDPL 798
KR HVFDE+ P TPLYN+KAYLPVI S F+ +LRA T QAFPQ VFDHWDM+ SDPL
Sbjct: 182 KRVHVFDEMHMPCTPLYNIKAYLPVI*SCGFSSTLRAATSCQAFPQCVFDHWDMMSSDPL 361
Query: 799 EPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
E G+ AA V +IRK+ GLKEQ+ PLSEFED+L
Sbjct: 362 EAGSQAAQLVTDIRKRNGLKEQMTPLSEFEDKL 460
>TC232891 similar to UP|Q9LNC5 (Q9LNC5) F9P14.8 protein, partial (26%)
Length = 785
Score = 197 bits (502), Expect = 1e-50
Identities = 101/259 (38%), Positives = 161/259 (61%), Gaps = 1/259 (0%)
Frame = +3
Query: 318 SALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDPEGPLMLYVSKMIPASDK 377
S +M++ H+PSP A +V+++Y GP D A+ CD GP+M+ V+K+ P SD
Sbjct: 12 SGFTDMLVQHIPSPRDAAIKKVDHIYAGPKDSSIYKAMAQCDSYGPVMVNVTKLYPKSDC 191
Query: 378 GRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRTVIWMGKKQETVEDVPCG 437
F AFGRV+SGK+ TG VR++G Y P +++D+ VK V + ++ + + V + P G
Sbjct: 192 SVFDAFGRVYSGKIQTGQTVRVLGEGYSPDDEEDMTVKEVTKLWVYQARDRMPVAEAPPG 371
Query: 438 NTVAMVGLDQFITKNATLTN-EKEVDAHPIRAMKFSVSPVVSVAVTCKVASDLPKLVEGL 496
+ V + G+D I K ATL N + + D + R ++F+ VV A S+LPK+VEGL
Sbjct: 372 SWVLIEGVDASIMKTATLCNVDYDEDVYIFRPLQFNTLSVVKTATEPLNPSELPKMVEGL 551
Query: 497 KRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNGAEITKSDPIVSFRETV 556
++++KS P+ V + E+GEH I GEL+L+ +KDL+ + + E+ +DP+VSF ETV
Sbjct: 552 RKISKSYPLAVTKVEESGEHTILGTGELYLDSIMKDLR-ELYSEVEVKVADPVVSFCETV 728
Query: 557 LEKSSHTVMSKSPNKHNRL 575
+E SS +++PNK N++
Sbjct: 729 VESSSMKCFAETPNKKNKI 785
>TC216233 GB|CAA50573.1|402753|GMFUSA translation elongation factor EF-G
{Glycine max;} , complete
Length = 2424
Score = 130 bits (327), Expect = 2e-30
Identities = 142/564 (25%), Positives = 235/564 (41%), Gaps = 15/564 (2%)
Frame = +1
Query: 8 RNMSVIAHVDHGKSTLTDSLVAAAGIIAQ--EVAGDVRMTDTRQDEAERGITIKSTGISL 65
RN+ ++AH+D GK+T T+ ++ G + EV D + E ERGITI S +
Sbjct: 49 RNIGIMAHIDAGKTTTTERILYYTGRNYKIGEVHEGTATMDWMEQEQERGITITSAATTT 228
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 125
++ NK+ IN+ID+PGHVDF+ EV ALR+ DGA+ + D V GV
Sbjct: 229 FW----------------NKHRINIIDTPGHVDFTLEVERALRVLDGAICLFDSVAGVEP 360
Query: 126 QTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYEDALL 185
Q+ETV RQA + + VNKMDR + + + + + + + + ED
Sbjct: 361 QSETVWRQADKYGVPRICFVNKMDRLGANFYRTRDMIVTNLGAKPLVIQLPIGS-EDNFK 537
Query: 186 GDVQVYPEKGTVSFSAGLHGWSFTLTNF------------AKMYASKFGVDEEKMMNRLW 233
G + + K V +S G F + + A+M + D++ M N L
Sbjct: 538 GVIDLVRNKAIV-WSGEELGAKFDIVDIPEDLQEQAQEYRAQMIENIVEFDDQAMENYLE 714
Query: 234 GENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNL 293
G + + KK ++G + + P+
Sbjct: 715 GIEPDEETIKK---------LIRKGTISASFVPV-------------------------- 789
Query: 294 KSEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYAS 353
+ G A + +Q LL+ ++ +LPSP + + ++P +
Sbjct: 790 ------MCGSAFKNKGVQ-------PLLDAVVDYLPSPLDLPAMKGSD-----PENPEET 915
Query: 354 AIRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLY 413
R + P K++ G F RV++GK+S G V N G+K
Sbjct: 916 IERVASDDEPFAGLAFKIMSDPFVGSL-TFVRVYAGKLSAGSYVL----NANKGKK---- 1068
Query: 414 VKSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSV 473
+ + R + +E V+ G+ +A+ GL IT + + + M F
Sbjct: 1069-ERIGRLLEMHANSREDVKVALAGDIIALAGLKDTITGETLCDPDNPI---VLERMDFP- 1233
Query: 474 SPVVSVAVTCKVASDLPKLVEGLKRLAKSDPMV-VCTISETGEHIIAAAGELHLEICLKD 532
PV+ VA+ K +D+ K+ GL +LA+ DP E + +I GELHLEI +
Sbjct: 1234DPVIKVAIEPKTKADVDKMATGLIKLAQEDPSFHFSRDEEINQTVIEGMGELHLEIIVDR 1413
Query: 533 LQDDFMNGAEITKSDPIVSFRETV 556
L+ +F E P V++RE++
Sbjct: 1414LKREFK--VEANVGAPQVNYRESI 1479
Score = 62.0 bits (149), Expect = 1e-09
Identities = 51/166 (30%), Positives = 76/166 (45%), Gaps = 7/166 (4%)
Frame = +2
Query: 636 VDTCKGVQYLNEIKDSVVAGFQIASKEGPMADENLRGVC--FEVCDV--VLHTDAIHRGG 691
+D G ++ +EIK V I + + GV F V DV VL + H
Sbjct: 1568 MDPGSGYEFKSEIKGGAVPKEYIPGVMKGLEECMSNGVLAGFPVVDVRAVLTDGSYHDVD 1747
Query: 692 GQIIP---TARRVFYAAMLTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQ 748
++ AR F + A PR+LEP+ VE+ PE LG + LN +RG +
Sbjct: 1748 SSVLAFQLAARGAFREGIRKAGPRMLEPIMKVEVVTPEEHLGDVIGDLNSRRGQINSFGD 1927
Query: 749 RPNTPLYNVKAYLPVIESFQFNESLRAQTGGQAFPQLVFDHWDMVP 794
+P L V A +P+ E FQ+ +LR T G+A + +D+VP
Sbjct: 1928 KPG-GLKVVDALVPLAEMFQYVSTLRGMTKGRASYTMQLAMFDVVP 2062
>TC230334 similar to UP|Q8RWP3 (Q8RWP3) GTP-binding protein LepA-like
protein, partial (27%)
Length = 820
Score = 101 bits (252), Expect = 1e-21
Identities = 60/144 (41%), Positives = 79/144 (54%)
Frame = +2
Query: 6 NIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISL 65
NIRN +IAH+DHGKSTL D L+ G + Q D + D E ERGITIK +
Sbjct: 311 NIRNFCIIAHIDHGKSTLADKLLQVTGTVHQREMKD-QFLDNMDLERERGITIKLQAARM 487
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 125
Y E Y +NLID+PGHVDFS EV+ +L +GAL+VVD +GV
Sbjct: 488 RYVF------------ENEPYCLNLIDTPGHVDFSYEVSRSLAACEGALLVVDASQGVEA 631
Query: 126 QTETVLRQALGERIKPVLTVNKMD 149
QT + AL ++ + +NK+D
Sbjct: 632 QTLANVYLALENNLEIIPVLNKID 703
>TC220357 homologue to UP|Q6SYB1 (Q6SYB1) GTP-binding protein TypA, partial
(25%)
Length = 668
Score = 98.6 bits (244), Expect = 1e-20
Identities = 58/145 (40%), Positives = 82/145 (56%)
Frame = +2
Query: 6 NIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISL 65
++RN++++AHVDHGK+TL D+++ + R+ D+ E ERGITI S S+
Sbjct: 227 DVRNIAIVAHVDHGKTTLVDAMLKQTKVFRDNQFVQERIMDSNDLERERGITILSKNTSV 406
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 125
Y+ D K IN+ID+PGH DF EV L + +G L+VVD VEG
Sbjct: 407 TYK----DAK------------INIIDTPGHSDFGGEVERILNMVEGILLVVDSVEGPMP 538
Query: 126 QTETVLRQALGERIKPVLTVNKMDR 150
QT VL++AL V+ VNK+DR
Sbjct: 539 QTRFVLKKALEFGHSVVVVVNKIDR 613
>TC231168 homologue to UP|Q9LNC5 (Q9LNC5) F9P14.8 protein, partial (13%)
Length = 697
Score = 92.0 bits (227), Expect = 9e-19
Identities = 50/113 (44%), Positives = 71/113 (62%), Gaps = 12/113 (10%)
Frame = +2
Query: 729 LGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQAFPQLVFD 788
+ IY+VL+++RGHV ++ +P TP Y VKA+LPVIESF F LR T GQAF VFD
Sbjct: 11 VSAIYTVLSRRRGHVTADVPQPGTPAYIVKAFLPVIESFGFETDLRYHTQGQAFCMSVFD 190
Query: 789 HWDMVPSDPLE------PGTPAAAR------VVEIRKKKGLKEQLIPLSEFED 829
HW +VP DPL+ P PA + +V+ R++KG+ E + +++F D
Sbjct: 191 HWAIVPGDPLDKSIVLRPLEPAPIQHLAREFMVKTRRRKGMSED-VSINKFFD 346
>AW707054 similar to GP|15450763|gb| elongation factor EF-2 {Arabidopsis
thaliana}, partial (7%)
Length = 193
Score = 83.2 bits (204), Expect = 4e-16
Identities = 41/52 (78%), Positives = 45/52 (85%)
Frame = +2
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEA 52
MD HNIRNMSVIAHVDH KSTLTDSLVAAAGIIAQE+ ++RMTDT D+A
Sbjct: 35 MDYNHNIRNMSVIAHVDHRKSTLTDSLVAAAGIIAQEITENMRMTDTHTDKA 190
>TC224434 similar to UP|Q9LS91 (Q9LS91) Elongation factor EF-2, partial (16%)
Length = 632
Score = 74.3 bits (181), Expect = 2e-13
Identities = 36/93 (38%), Positives = 51/93 (54%)
Frame = +1
Query: 705 AMLTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVI 764
A++ PR +E +Y E+ LG +Y+VL++ V E + +P + V AYLPV
Sbjct: 1 AVVQXXPRXVEAMYFCELNTXTEYLGPMYAVLSRXXARVLKEEMQEGSPFFTVHAYLPVX 180
Query: 765 ESFQFNESLRAQTGGQAFPQLVFDHWDMVPSDP 797
ESF F + LR T G A LV HW+ +P DP
Sbjct: 181 ESFGFADELRRWTSGAASALLVLSHWEALPEDP 279
>TC204394 similar to UP|EF2_BETVU (O23755) Elongation factor 2 (EF-2),
partial (5%)
Length = 497
Score = 70.9 bits (172), Expect = 2e-12
Identities = 32/49 (65%), Positives = 37/49 (75%)
Frame = +3
Query: 614 WDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIASKE 662
++ +LAKK+WCFGPET GPNM+VD CKGVQYLNEI VAG KE
Sbjct: 348 FESNLAKKIWCFGPETLGPNMVVDMCKGVQYLNEIXGFWVAGXSGHXKE 494
>TC214026 similar to UP|Q6G0R7 (Q6G0R7) GTP-binding protein typA, partial
(14%)
Length = 501
Score = 63.9 bits (154), Expect = 3e-10
Identities = 38/108 (35%), Positives = 57/108 (52%)
Frame = +1
Query: 7 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLY 66
+RN++VIAHVDHGK+TL D L+ G R D+ E ERGITI S S+
Sbjct: 238 LRNLAVIAHVDHGKTTLMDRLLRQCGADLPH----ERAMDSISLERERGITISSKVTSVS 405
Query: 67 YEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGAL 114
++ ++ +N++D+PGH DF EV + + +GA+
Sbjct: 406 WKENE----------------LNMVDTPGHADFGGEVERVVGMVEGAI 501
>BF068251 similar to GP|15450763|gb| elongation factor EF-2 {Arabidopsis
thaliana}, partial (5%)
Length = 139
Score = 58.9 bits (141), Expect = 9e-09
Identities = 27/37 (72%), Positives = 33/37 (88%)
Frame = +2
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQE 37
+D KHN +N+SVIAHVDH KSTLTDS+VAAAG+IA +
Sbjct: 29 IDYKHNSQNVSVIAHVDHRKSTLTDSIVAAAGVIAHQ 139
>TC214319 homologue to UP|EFT1_SOYBN (Q43467) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 1583
Score = 58.2 bits (139), Expect = 2e-08
Identities = 50/175 (28%), Positives = 76/175 (42%), Gaps = 2/175 (1%)
Frame = +1
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYE 68
N+ I HVDHGK+TLT +L A + D +E RGITI + +
Sbjct: 250 NIGTIGHVDHGKTTLTAALTMALAALGNSAPKKYDEIDAAPEERARGITINTATVEY--- 420
Query: 69 MSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTE 128
E E Y +D PGH D+ + DGA++VV +G QT+
Sbjct: 421 -----------ETENRHYA--HVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTK 561
Query: 129 --TVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYE 181
+L + +G V+ +NK D+ +D EE +Q V V ++++YE
Sbjct: 562 EHILLAKQVGVP-NMVVFLNKQDQ------VDDEE---LLQLVELEVRDLLSSYE 696
>TC214405 homologue to UP|EFT1_SOYBN (Q43467) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 2050
Score = 57.8 bits (138), Expect = 2e-08
Identities = 50/175 (28%), Positives = 75/175 (42%), Gaps = 2/175 (1%)
Frame = +3
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYE 68
N+ I HVDHGK+TLT +L A + D +E RGITI + +
Sbjct: 273 NIGTIGHVDHGKTTLTAALTMALAALGNSAPKKYDEIDAAPEERARGITINTATVEY--- 443
Query: 69 MSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTE 128
E E Y +D PGH D+ + DGA++VV +G QT+
Sbjct: 444 -----------ETENRHYA--HVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTK 584
Query: 129 --TVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYE 181
+L + +G V+ +NK D+ +D EE +Q V V ++ +YE
Sbjct: 585 EHILLAKQVGVP-NMVVFLNKQDQ------VDDEE---LLQLVELEVRDLLTSYE 719
>TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 1615
Score = 56.2 bits (134), Expect = 6e-08
Identities = 40/143 (27%), Positives = 60/143 (40%), Gaps = 1/143 (0%)
Frame = +1
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYE 68
N+ I HVDHGK+TLT +L A + D +E RGITI + +
Sbjct: 250 NIGTIGHVDHGKTTLTAALTMALASLGNSAPKKYDEIDAAPEERARGITINTATVEY--- 420
Query: 69 MSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTE 128
E E Y +D PGH D+ + DGA++VV +G QT+
Sbjct: 421 -----------ETENRHYA--HVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTK 561
Query: 129 TVLRQALGERIKPVLT-VNKMDR 150
+ A + ++ +NK D+
Sbjct: 562 EHILLAKQVGVPNIVVFLNKQDQ 630
>TC217724 homologue to UP|EFTM_ARATH (Q9ZT91) Elongation factor Tu,
mitochondrial precursor, partial (87%)
Length = 1679
Score = 52.4 bits (124), Expect = 8e-07
Identities = 39/122 (31%), Positives = 56/122 (44%), Gaps = 2/122 (1%)
Frame = +2
Query: 9 NMSVIAHVDHGKSTLTDSL--VAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLY 66
N+ I HVDHGK+TLT ++ V A + A+ VA D D +E +RGITI + +
Sbjct: 308 NVGTIGHVDHGKTTLTAAITRVLADEVKAKAVAFD--EIDKAPEEKKRGITIATAHV--- 472
Query: 67 YEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQ 126
E E K +D PGH D+ + DG ++VV +G Q
Sbjct: 473 -------------EYETAKRHYAHVDCPGHADYVKNMITGAAQMDGGILVVSAPDGPMPQ 613
Query: 127 TE 128
T+
Sbjct: 614 TK 619
>AI442962 weakly similar to GP|10798636|emb elongation factor 2 {Nicotiana
tabacum}, partial (38%)
Length = 217
Score = 50.1 bits (118), Expect = 4e-06
Identities = 30/72 (41%), Positives = 37/72 (50%)
Frame = -1
Query: 748 QRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAAR 807
+R P K PV ES+ +L GQAFPQ VFDH + SDPL+ + A
Sbjct: 217 RRLGPPF*QSKTTFPVNESWCSQGTLGGAKKGQAFPQCVFDHGKKMSSDPLKGESQMAKL 38
Query: 808 VVEIRKKKGLKE 819
V K+KGLKE
Sbjct: 37 VXHTPKRKGLKE 2
>TC203954 homologue to UP|EF1A_LYCES (P17786) Elongation factor 1-alpha
(EF-1-alpha), partial (61%)
Length = 1423
Score = 47.8 bits (112), Expect = 2e-05
Identities = 40/129 (31%), Positives = 57/129 (44%), Gaps = 15/129 (11%)
Frame = +3
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVR--------MTDTRQDEAE 53
N+ VI HVD GKST T L+ G I + V A ++ + D + E E
Sbjct: 456 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 635
Query: 54 RGITIKSTGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGA 113
RGITI I+L+ + E KY +ID+PGH DF + D A
Sbjct: 636 RGITI---DIALW-------------KFETTKYYCTVIDAPGHRDFIKNMITGTSQADCA 767
Query: 114 LVVVDCVEG 122
++++D G
Sbjct: 768 VLIIDSTTG 794
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,302,043
Number of Sequences: 63676
Number of extensions: 413020
Number of successful extensions: 1932
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 1908
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1919
length of query: 831
length of database: 12,639,632
effective HSP length: 105
effective length of query: 726
effective length of database: 5,953,652
effective search space: 4322351352
effective search space used: 4322351352
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146817.8


