

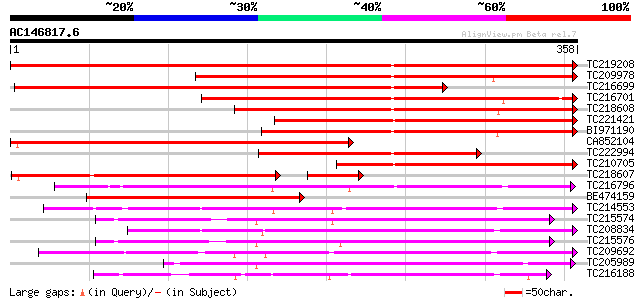
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146817.6 - phase: 0
(358 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219208 weakly similar to UP|Q948K9 (Q948K9) CmE8 protein, part... 541 e-154
TC209978 weakly similar to GB|AAC49826.1|1916643|CRU71604 desace... 298 3e-81
TC216699 weakly similar to UP|Q84MB3 (Q84MB3) At1g06620, partial... 296 1e-80
TC216701 weakly similar to UP|Q84MB3 (Q84MB3) At1g06620, partial... 263 7e-71
TC218608 weakly similar to UP|Q9LTH8 (Q9LTH8) Leucoanthocyanidin... 256 8e-69
TC221421 weakly similar to GB|AAC49826.1|1916643|CRU71604 desace... 255 2e-68
BI971190 224 6e-59
CA852104 224 6e-59
TC222994 weakly similar to UP|Q84MB3 (Q84MB3) At1g06620, partial... 210 9e-55
TC210705 weakly similar to UP|Q9LSW7 (Q9LSW7) 1-aminocyclopropan... 200 7e-52
TC218607 weakly similar to UP|ACC3_LYCES (P10967) 1-aminocyclopr... 182 9e-51
TC216796 similar to GB|AAQ65160.1|34365697|BT010537 At4g10500 {A... 181 6e-46
BE474159 weakly similar to GP|8885558|dbj leucoanthocyanidin dio... 169 2e-42
TC214553 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 166 1e-41
TC215574 similar to UP|O04076 (O04076) ACC-oxidase, partial (98%) 166 2e-41
TC208834 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 163 1e-40
TC215576 similar to UP|O04076 (O04076) ACC-oxidase, partial (97%) 160 1e-39
TC209692 weakly similar to UP|Q39224 (Q39224) SRG1 protein (F6I1... 157 5e-39
TC205989 similar to UP|Q9FLV0 (Q9FLV0) Flavanone 3-hydroxylase-l... 156 2e-38
TC216188 similar to UP|Q84L58 (Q84L58) 1-aminocyclopropane-1-car... 155 3e-38
>TC219208 weakly similar to UP|Q948K9 (Q948K9) CmE8 protein, partial (87%)
Length = 1242
Score = 541 bits (1395), Expect = e-154
Identities = 252/358 (70%), Positives = 301/358 (83%)
Frame = +1
Query: 1 MDSNRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFA 60
++ +RL ELKAFDDTK GVKGLVD+GV KIP LFHHP D+F A+ NT++ IP+ID A
Sbjct: 61 LNPDRLGELKAFDDTKAGVKGLVDEGVAKIPRLFHHPPDEFVMASKLGNTEYTIPVIDLA 240
Query: 61 NFGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELY 120
G DP +R EI +IREA E WGFFQVVNHGIP+ VLED+++GV RF EQD+E KKELY
Sbjct: 241 EVGKDPSSRQEIIGRIREASERWGFFQVVNHGIPVTVLEDLKDGVQRFHEQDIEEKKELY 420
Query: 121 SRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKL 180
+RD M+P YNSNF+LYSS ALNWRD+F+C LAPN PKP+DLP+VCRD LLEYG YV KL
Sbjct: 421 TRDHMKPLVYNSNFDLYSSPALNWRDSFMCYLAPNPPKPEDLPVVCRDILLEYGTYVMKL 600
Query: 181 GMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLL 240
G+ LFELLSEALGLHP+HLKD+GCA+G++SL HYYPACPEP+LT+GTTKHSD+CFLT+LL
Sbjct: 601 GIALFELLSEALGLHPDHLKDLGCAEGLISLCHYYPACPEPDLTLGTTKHSDNCFLTVLL 780
Query: 241 QDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSV 300
Q DHIGGLQV +QN WIDI P P ALVVN+GDLLQL+TND+FKSVEH+V AN+IGPR+SV
Sbjct: 781 Q-DHIGGLQVLYQNMWIDITPEPGALVVNIGDLLQLITNDRFKSVEHRVQANLIGPRISV 957
Query: 301 ACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
ACFF+ L S YGPIKE L+EDNPPKY+E T++EYV Y+ KG+DG SALQHF++
Sbjct: 958 ACFFSEGLKSSPKPYGPIKELLTEDNPPKYRETTVAEYVRYFEAKGLDGTSALQHFKI 1131
>TC209978 weakly similar to GB|AAC49826.1|1916643|CRU71604
desacetoxyvindoline 4-hydroxylase {Catharanthus roseus;}
, partial (48%)
Length = 864
Score = 298 bits (762), Expect = 3e-81
Identities = 142/244 (58%), Positives = 182/244 (74%), Gaps = 3/244 (1%)
Frame = +3
Query: 118 ELYSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYV 177
E YSRD + SY SN+NLY + NWRDTF +AP+ PKP+++ VCRD ++EY + +
Sbjct: 3 EFYSRDIKKKVSYYSNYNLYHDNPANWRDTFGFVVAPDPPKPEEISSVCRDIVIEYSKKI 182
Query: 178 TKLGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLT 237
LG T+FELLSEALGL+P++LK+ C +G+ L HYYP CPEP LTMGTTKH+DS F+T
Sbjct: 183 RDLGFTIFELLSEALGLNPSYLKEFNCGEGLFILGHYYPTCPEPGLTMGTTKHTDSNFMT 362
Query: 238 LLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPR 297
LLLQ D IGGLQV HQN+W+++PP+ ALVVN+GDLLQL+TNDKF SV H+VL+ GPR
Sbjct: 363 LLLQ-DQIGGLQVLHQNQWVNVPPLHGALVVNIGDLLQLITNDKFVSVYHRVLSQNTGPR 539
Query: 298 VSVACFF---NADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQ 354
+SVA FF + S YGPIKE LSE+NPP Y++ T+ E++AYY KG+DG S+L
Sbjct: 540 ISVASFFVNSHDPAEGTSKMYGPIKELLSEENPPIYRDTTLKEFLAYYYAKGLDGNSSLD 719
Query: 355 HFRV 358
FRV
Sbjct: 720 PFRV 731
>TC216699 weakly similar to UP|Q84MB3 (Q84MB3) At1g06620, partial (53%)
Length = 1436
Score = 296 bits (758), Expect = 1e-80
Identities = 142/274 (51%), Positives = 192/274 (69%), Gaps = 1/274 (0%)
Frame = +1
Query: 4 NRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKA-TNSNNTQHIIPIIDFANF 62
+R E+KAFDD+KTGV+GLV+ GV K+P +F+ T ++ ++ IPIID
Sbjct: 286 DRKSEIKAFDDSKTGVQGLVENGVTKVPLMFYSENSNLNDGVTGASYSKISIPIIDLTGI 465
Query: 63 GNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSR 122
+DP R + K+R ACE WGFFQV+NHGIP +VL++M G RF +QD +V+KE Y+R
Sbjct: 466 HDDPILRDHVVGKVRYACEKWGFFQVINHGIPTHVLDEMIKGTCRFHQQDAKVRKEYYTR 645
Query: 123 DPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGM 182
+ R +Y SN+ L+ + +WRDT SLAP+ P+ ++ P VCRD + EY + + L
Sbjct: 646 EVSRKVAYLSNYTLFEDPSADWRDTLAFSLAPHPPEAEEFPAVCRDIVNEYSKKIMALAY 825
Query: 183 TLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQD 242
LFELLSEALGL+ +LK+M CA+G L L HYYPACPEPELTMG TKHSD +T+LLQ
Sbjct: 826 ALFELLSEALGLNRFYLKEMDCAEGQLLLCHYYPACPEPELTMGNTKHSDGNTITILLQ- 1002
Query: 243 DHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQL 276
D IGGLQV H ++WID+PP+ ALVVN+GD++Q+
Sbjct: 1003DQIGGLQVLHDSQWIDVPPMHGALVVNIGDIMQV 1104
>TC216701 weakly similar to UP|Q84MB3 (Q84MB3) At1g06620, partial (50%)
Length = 974
Score = 263 bits (673), Expect = 7e-71
Identities = 134/240 (55%), Positives = 169/240 (69%), Gaps = 3/240 (1%)
Frame = +3
Query: 122 RDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLG 181
RD R Y SNF+LY + +WRDT APN+P ++LP VCRD + EY V L
Sbjct: 3 RDMSRKVIYLSNFSLYQDPSADWRDTLAFFWAPNSPNDEELPAVCRDIVPEYSTKVMALA 182
Query: 182 MTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQ 241
TLFELLSEALGL HLK+MGC +G+L L HYYPACPEPELTMGT++H+D F+T+LLQ
Sbjct: 183 STLFELLSEALGLDRFHLKEMGCDEGLLHLCHYYPACPEPELTMGTSRHTDGNFMTILLQ 362
Query: 242 DDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVA 301
D +GGLQ+ H+N+WID+P ALVVN+GDLLQLVTN+KF SV+H+VLAN GPR S+A
Sbjct: 363 -DQMGGLQILHENQWIDVPAAHGALVVNIGDLLQLVTNEKFISVQHRVLANHQGPRTSIA 539
Query: 302 CFFNADLNS---FSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
FF S S +GPIKE LSE NPP Y+E ++ +Y+A+ K I G S+L F++
Sbjct: 540 SFFRIGDQSPEGLSRVFGPIKELLSEHNPPVYRETSLKDYLAHQYAKSI-GASSLSLFKL 716
>TC218608 weakly similar to UP|Q9LTH8 (Q9LTH8) Leucoanthocyanidin
dioxygenase-like protein (At5g59530), partial (57%)
Length = 806
Score = 256 bits (655), Expect = 8e-69
Identities = 124/219 (56%), Positives = 163/219 (73%), Gaps = 3/219 (1%)
Frame = +1
Query: 143 NWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGMTLFELLSEALGLHPNHLKDM 202
NWRD+ CS+APN PKP++LP V RD ++EY + + LG T+FELLSEALGL+P++LK+M
Sbjct: 34 NWRDSLGCSMAPNPPKPENLPTVFRDIIIEYSKEIMALGCTIFELLSEALGLNPSYLKEM 213
Query: 203 GCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIPPI 262
CA+ + HYYP CPEPELT+GT+KH+DS F+T++LQ +GGLQV H+ W ++PP+
Sbjct: 214 NCAERLFIQGHYYPPCPEPELTLGTSKHTDSSFMTIVLQ-GQLGGLQVLHEKLWFNVPPV 390
Query: 263 PDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNAD---LNSFSNQYGPIK 319
ALVVNVGD+LQL+TND F SV H+VL+N GPRVSVA FF+ S Y PIK
Sbjct: 391 HGALVVNVGDILQLITNDNFVSVYHRVLSNHGGPRVSVASFFSNSHDPAKGASMVYSPIK 570
Query: 320 EFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
E LSE+NP Y++ TI E +A++ KG+DG SALQ FR+
Sbjct: 571 ELLSEENPAIYRDTTIGEIMAHHFAKGLDGNSALQPFRL 687
>TC221421 weakly similar to GB|AAC49826.1|1916643|CRU71604
desacetoxyvindoline 4-hydroxylase {Catharanthus roseus;}
, partial (42%)
Length = 1072
Score = 255 bits (651), Expect = 2e-68
Identities = 123/191 (64%), Positives = 146/191 (76%)
Frame = +1
Query: 168 DELLEYGEYVTKLGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGT 227
D +EY V LG LF LLSEALGL P+HL+ M CAKG L HYYP+CPEPELTMGT
Sbjct: 73 DVAMEYSRQVKLLGRVLFGLLSEALGLDPDHLEGMDCAKGHSILFHYYPSCPEPELTMGT 252
Query: 228 TKHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEH 287
T+HSD FLT+LLQD HIGGLQV N W+D+PP+P ALVVN+GDLLQL++NDKFKSVEH
Sbjct: 253 TRHSDPDFLTILLQD-HIGGLQVLSHNGWVDVPPVPGALVVNIGDLLQLISNDKFKSVEH 429
Query: 288 KVLANIIGPRVSVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGI 347
+VLAN IGPRVSVACFF L + YGPIKE LSE+ PP Y+E ++ +++AYY K +
Sbjct: 430 RVLANRIGPRVSVACFFTIHLYPTTRIYGPIKELLSEEKPPVYRETSLKDFIAYYYNKEL 609
Query: 348 DGISALQHFRV 358
DG SAL HF +
Sbjct: 610 DGNSALDHFMI 642
>BI971190
Length = 772
Score = 224 bits (570), Expect = 6e-59
Identities = 107/206 (51%), Positives = 145/206 (69%), Gaps = 7/206 (3%)
Frame = -2
Query: 160 QDLPLVCRDELLEYGEYVTKLGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACP 219
+ +P VC D ++EY + +G T+FELLSEALG + ++L ++ C +G+ ++ +YYPACP
Sbjct: 759 EHIPSVCXDIVIEYTKXXRDVGFTIFELLSEALGXNSSYLNELSCGEGLFTVGNYYPACP 580
Query: 220 EPELTMGTTKHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTN 279
EPELTMG KH+D F+TLLLQ D IGGLQV HQN+W+++PP+ ALV VGDLLQL+TN
Sbjct: 579 EPELTMGAAKHTDGNFMTLLLQ-DQIGGLQVLHQNQWVNLPPVHGALVXXVGDLLQLITN 403
Query: 280 DKFKSVEHKVLANIIGPRVSVACFFNA-------DLNSFSNQYGPIKEFLSEDNPPKYKE 332
DKF SV H+VL+ PR+SVA FF + YGPIKE +SE+NPP Y++
Sbjct: 402 DKFVSVCHRVLSKKTCPRISVASFFGTFFGHSDDPVEGLQKLYGPIKELISEENPPIYRD 223
Query: 333 ITISEYVAYYNTKGIDGISALQHFRV 358
TI ++VAYY K +DG S+L FR+
Sbjct: 222 TTIKDFVAYYYAKALDGKSSLNRFRL 145
>CA852104
Length = 723
Score = 224 bits (570), Expect = 6e-59
Identities = 107/220 (48%), Positives = 155/220 (69%), Gaps = 3/220 (1%)
Frame = +2
Query: 1 MDS--NRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHH-PLDKFGKATNSNNTQHIIPII 57
MDS +R E+KAFDD+K GVKGLV+ GV KIP +FH LD T+ +++ IIPII
Sbjct: 62 MDSTYDRKAEVKAFDDSKAGVKGLVESGVTKIPRMFHSGKLDLDIIETSGGDSKLIIPII 241
Query: 58 DFANFGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKK 117
DF + ++P R E+ IR AC WGFFQV+NHGIP++VL++M +G+ RF EQD E +K
Sbjct: 242 DFKDIHSNPALRSEVIGTIRSACHEWGFFQVINHGIPISVLDEMIDGIRRFHEQDTEARK 421
Query: 118 ELYSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYV 177
E Y+RD + Y SN L+S + +NWRDT +++P+ PKP+ +P VCRD ++EY + +
Sbjct: 422 EFYTRDSKKKVVYFSNLGLHSGNPVNWRDTIGFAVSPDPPKPEHIPSVCRDIVIEYTKKI 601
Query: 178 TKLGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPA 217
+G T+FELLSEALGL+ ++L ++ C +G+ ++ +YYPA
Sbjct: 602 RDVGFTIFELLSEALGLNSSYLNELSCGEGLFTVGNYYPA 721
>TC222994 weakly similar to UP|Q84MB3 (Q84MB3) At1g06620, partial (38%)
Length = 421
Score = 210 bits (534), Expect = 9e-55
Identities = 96/141 (68%), Positives = 120/141 (85%)
Frame = +2
Query: 158 KPQDLPLVCRDELLEYGEYVTKLGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPA 217
KP+DLP VCRD LLEY + V KLG LFELLSEAL L+PN+L D+GC +G+ + H YPA
Sbjct: 2 KPEDLPSVCRDILLEYAKEVKKLGSVLFELLSEALELNPNYLNDIGCNEGLTLVCHCYPA 181
Query: 218 CPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLV 277
CPEPELT+G TKH+D+ F+T+LLQ DHIGGLQV H+N+W+D+ P+P ALV+N+GDLLQL+
Sbjct: 182 CPEPELTLGATKHTDNDFITVLLQ-DHIGGLQVLHENRWVDVSPVPGALVINIGDLLQLI 358
Query: 278 TNDKFKSVEHKVLANIIGPRV 298
TNDKFKSVEH+V+AN +GPRV
Sbjct: 359 TNDKFKSVEHRVVANRVGPRV 421
>TC210705 weakly similar to UP|Q9LSW7 (Q9LSW7)
1-aminocyclopropane-1-carboxylate oxidase
(AT5g43440/MWF20_15), partial (35%)
Length = 510
Score = 200 bits (509), Expect = 7e-52
Identities = 91/152 (59%), Positives = 116/152 (75%)
Frame = +2
Query: 207 GILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIPPIPDAL 266
G + HYYP+CPE ELT+GT KH+D F+T+LLQD HIGGLQV HQ+ WID+ P+P AL
Sbjct: 2 GQFAFGHYYPSCPESELTLGTIKHADVDFITVLLQD-HIGGLQVLHQDTWIDVTPVPGAL 178
Query: 267 VVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNADLNSFSNQYGPIKEFLSEDN 326
VVN+GD LQL++NDKFKS +H+VLA +GPRVS+ACFF+ + S Y PIKE LSEDN
Sbjct: 179 VVNIGDFLQLISNDKFKSAQHRVLAKTVGPRVSIACFFSPAFHPSSRTYAPIKELLSEDN 358
Query: 327 PPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
P KY+E +I E+ A+Y TK + G S L HF++
Sbjct: 359 PAKYREFSIPEFTAHYRTKCMKGTSPLLHFKI 454
>TC218607 weakly similar to UP|ACC3_LYCES (P10967)
1-aminocyclopropane-1-carboxylate oxidase homolog
(Protein E8), partial (35%)
Length = 860
Score = 182 bits (463), Expect(2) = 9e-51
Identities = 87/172 (50%), Positives = 119/172 (68%), Gaps = 2/172 (1%)
Frame = +1
Query: 2 DSN--RLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDF 59
DSN R E+KAFDD+KTGVKGLVD GV KIP +FH ++ + +N IP+ID
Sbjct: 133 DSNYDRKAEIKAFDDSKTGVKGLVDSGVKKIPRMFHSGINITENVASDSNLS--IPVIDL 306
Query: 60 ANFGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKEL 119
+ N+P +E+ +KIR AC+ WGFFQV+NHGIP++V++ M +G+ RF EQD +V+K+
Sbjct: 307 QDIHNNPALHNEVVTKIRSACQEWGFFQVINHGIPISVMDQMIDGIRRFHEQDTDVRKQF 486
Query: 120 YSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELL 171
YSRD + YNSN +LY NWRD+ CS+APN PKP++LP V RD ++
Sbjct: 487 YSRDLKKTILYNSNTSLYLDKFANWRDSLGCSMAPNPPKPENLPTVFRDIII 642
Score = 35.8 bits (81), Expect(2) = 9e-51
Identities = 18/36 (50%), Positives = 23/36 (63%), Gaps = 1/36 (2%)
Frame = +3
Query: 189 SEALGLHPNHLKDMGCAKGILSLS-HYYPACPEPEL 223
SEALG +P++LK G L + HYYP CP+ EL
Sbjct: 699 SEALGPNPSYLKGNGIGAXRLFIQGHYYPPCPDLEL 806
>TC216796 similar to GB|AAQ65160.1|34365697|BT010537 At4g10500 {Arabidopsis
thaliana;} , partial (72%)
Length = 1454
Score = 181 bits (458), Expect = 6e-46
Identities = 110/338 (32%), Positives = 177/338 (51%), Gaps = 9/338 (2%)
Frame = +1
Query: 29 KIPTLFHHPL-DKFGKATNSNNTQHIIPIIDFANFGNDPKTRHEITSKIREACETWGFFQ 87
++P+ F PL D+ ++ IP+ID + + P H I +I +AC+ +GFFQ
Sbjct: 136 QVPSNFIRPLGDRPNLQGVVQSSDVCIPLIDLQDL-HGPNRSH-IIQQIDQACQNYGFFQ 309
Query: 88 VVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRPFSYNSNFNLYSSSALNWRDT 147
V NHG+P V+E + FF K + YS DP + +++FN+ S +WRD
Sbjct: 310 VTNHGVPEGVIEKIMKVTREFFGLPESEKLKSYSTDPFKASRLSTSFNVNSEKVSSWRDF 489
Query: 148 FVCSLAPNAPKPQDLPL---VCRDELLEYGEYVTKLGMTLFELLSEALGLHPNHLKDMGC 204
P ++ P R+++ EY + + + L E +SE+LGL +++ +
Sbjct: 490 LRLHCHPIEDYIKEWPSNPPSLREDVAEYCRKMRGVSLKLVEAISESLGLERDYINRVVG 669
Query: 205 AKGILSLSH----YYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIP 260
K H YYPACPEPELT G H+D +T+LLQD+ + GLQV KW+ +
Sbjct: 670 GKKGQEQQHLAMNYYPACPEPELTYGLPGHTDPTVITILLQDE-VPGLQVLKDGKWVAVN 846
Query: 261 PIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNADLNSFSNQYGPIKE 320
PIP+ VVNVGD +Q+++NDK+KSV H+ + N R+S+ F+ ++ GP +
Sbjct: 847 PIPNTFVVNVGDQIQVISNDKYKSVLHRAVVNCNKDRISIPTFYFPSNDAI---IGPAPQ 1017
Query: 321 FL-SEDNPPKYKEITISEYVAYYNTKGIDGISALQHFR 357
+ +PP+Y T +EY + +G+ + L F+
Sbjct: 1018LIHHHHHPPQYNNFTYNEYYQNFWNRGLSKETCLDIFK 1131
>BE474159 weakly similar to GP|8885558|dbj leucoanthocyanidin
dioxygenase-like protein {Arabidopsis thaliana}, partial
(30%)
Length = 426
Score = 169 bits (428), Expect = 2e-42
Identities = 78/139 (56%), Positives = 101/139 (72%), Gaps = 1/139 (0%)
Frame = +3
Query: 49 NTQHIIPIIDFANFGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRF 108
N H+IPIID A+ DP R + +++A ETWGFFQV+NH IPL+VLE+M+NGV RF
Sbjct: 9 NKSHVIPIIDLADIDKDPSKRQGLVDIVKKASETWGFFQVINHDIPLSVLEEMKNGVKRF 188
Query: 109 FEQDVEVKKELYSRDPMRPFSYNSNFNLYSSS-ALNWRDTFVCSLAPNAPKPQDLPLVCR 167
E D E KKE YSRD + F YNSNF+LY S A+NWRD+ C L P+ PKP++LP+VCR
Sbjct: 189 HEMDTEAKKEFYSRDRSKSFLYNSNFDLYGSQPAINWRDSCRCLLYPDTPKPEELPVVCR 368
Query: 168 DELLEYGEYVTKLGMTLFE 186
D LLEY +++ +LG+ L E
Sbjct: 369 DILLEYRKHIMRLGILLLE 425
>TC214553 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (52%)
Length = 1808
Score = 166 bits (421), Expect = 1e-41
Identities = 101/343 (29%), Positives = 181/343 (52%), Gaps = 6/343 (1%)
Frame = +1
Query: 22 LVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFANFGNDPKTRHEITSKIREACE 81
L + ++++P + H +N+ + + PIID ++ + E K+ AC+
Sbjct: 73 LAKQPIIEVPERYVHANQDPHILSNTISLPQV-PIIDLHQLLSEDPSELE---KLDHACK 240
Query: 82 TWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRPFSYNSNFNLYSSSA 141
WGFFQ++NHG+ V+E+M+ GV FF +E K++ + + P + F +
Sbjct: 241 EWGFFQLINHGVDPPVVENMKIGVQEFFNLPMEEKQKFW-QTPEDMQGFGQLFVVSEEQK 417
Query: 142 LNWRDTFVCSLAP-NAPKPQDLPLV---CRDELLEYGEYVTKLGMTLFELLSEALGLHPN 197
L W D F P ++ P +P + R+ L Y + K+ +T+ L+ +AL + N
Sbjct: 418 LEWADMFYAHTFPLHSRNPHLIPKIPQPFRENLENYCLELRKMCITIIGLMKKALKIKTN 597
Query: 198 HLKDM--GCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQVFHQNK 255
L ++ ++GI +YYP CP+PE +G HSDS LT+LLQ + + GLQ+ K
Sbjct: 598 ELSELFEDPSQGIRM--NYYPPCPQPERVIGINPHSDSGALTILLQVNEVEGLQIRKDGK 771
Query: 256 WIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNADLNSFSNQY 315
WI + P+ +A V+NVGD+L+++TN ++S+EH+ + N R+S+A F + S
Sbjct: 772 WIPVKPLSNAFVINVGDMLEILTNGIYRSIEHRGIVNSEKERISIAMFHRPQM---SRVI 942
Query: 316 GPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
GP ++ + P +K I +++Y+ + + + G S + R+
Sbjct: 943 GPAPSLVTPERPALFKRIGVADYLNGFLKRELKGKSYMDVIRI 1071
>TC215574 similar to UP|O04076 (O04076) ACC-oxidase, partial (98%)
Length = 1169
Score = 166 bits (420), Expect = 2e-41
Identities = 96/296 (32%), Positives = 154/296 (51%), Gaps = 6/296 (2%)
Frame = +3
Query: 55 PIIDFANFGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVE 114
P+I+ N + R ++I +AC+ WGFF++VNHGIPL +L+ + + + +E
Sbjct: 51 PVINLENLNGEE--RKATLNQIEDACQNWGFFELVNHGIPLELLDTVERLTKEHYRKCME 224
Query: 115 VK-KELYSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAP--NAPKPQDLPLVCRDELL 171
+ KE S + ++W TF P N + DL RD +
Sbjct: 225 KRFKEAVSSKGLEA----------EVKDMDWESTFFLRHLPTSNISEIPDLSQEYRDAMK 374
Query: 172 EYGEYVTKLGMTLFELLSEALGLHPNHLKDM---GCAKGILSLSHYYPACPEPELTMGTT 228
E+ + + KL L +LL E LGL +LK+ + YPACP+PEL G
Sbjct: 375 EFAQKLEKLAEELLDLLCENLGLEKGYLKNAFYGSRGPNFGTKVANYPACPKPELVKGLR 554
Query: 229 KHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHK 288
H+D+ + LLLQDD + GLQ+ +W+D+PP+ ++VVN+GD ++++TN ++KSVEH+
Sbjct: 555 AHTDAGGIILLLQDDKVSGLQLLKNGQWVDVPPMRHSIVVNLGDQIEVITNGRYKSVEHR 734
Query: 289 VLANIIGPRVSVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNT 344
V+A G R+SVA F+N ++ + E +ED Y + +Y+ Y T
Sbjct: 735 VIAQTNGTRMSVASFYNPASDALIYPAPALLEQKAEDTEQVYPKFVFEDYMKLYAT 902
>TC208834 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (54%)
Length = 1070
Score = 163 bits (413), Expect = 1e-40
Identities = 89/289 (30%), Positives = 152/289 (51%), Gaps = 5/289 (1%)
Frame = +2
Query: 75 KIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRPFSYNSNF 134
K+ AC+ WGFFQ+VNHG+ +++E +R FF + KK+ + + P + F
Sbjct: 32 KLHLACKEWGFFQLVNHGVNSSLVEKVRLETQDFFNLPMSEKKKFW-QTPQHMEGFGQAF 208
Query: 135 NLYSSSALNWRDTFVCSLAPNAPK-----PQDLPLVCRDELLEYGEYVTKLGMTLFELLS 189
+ L+W D + + P + PQ LPL RD L Y + L + + L+
Sbjct: 209 VVSEDQKLDWADLYYMTTLPKHSRMPHLFPQ-LPLPFRDTLEAYSREIKDLAIVIIGLMG 385
Query: 190 EALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQ 249
+AL + ++++ L +YYP CPEPE +G T HSD L +LLQ + + GLQ
Sbjct: 386 KALKIQEREIRELFEDGIQLMRMNYYPPCPEPEKVIGLTPHSDGIGLAILLQLNEVEGLQ 565
Query: 250 VFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNADLN 309
+ W+ + P+ +A +VNVGD+L+++TN ++S+EH+ N R+S A F++
Sbjct: 566 IRKDGLWVPVKPLINAFIVNVGDILEIITNGIYRSIEHRATVNGEKERLSFATFYSP--- 736
Query: 310 SFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISALQHFRV 358
S GP ++E PP++K I + +Y ++ +DG + ++ R+
Sbjct: 737 SSDGVVGPAPSLITEQTPPRFKSIGVKDYFKGLFSRKLDGKAYIEVMRI 883
>TC215576 similar to UP|O04076 (O04076) ACC-oxidase, partial (97%)
Length = 1151
Score = 160 bits (404), Expect = 1e-39
Identities = 95/296 (32%), Positives = 157/296 (52%), Gaps = 6/296 (2%)
Frame = +2
Query: 55 PIIDFANFGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVE 114
P+I+ N + R +I +AC+ WGFF++VNHGIPL +L+ + + + +E
Sbjct: 2 PVINLDNLNGEE--RKATLDQIEDACQNWGFFELVNHGIPLELLDTVERLTKEHYRKCME 175
Query: 115 VK-KELYSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAP--NAPKPQDLPLVCRDELL 171
+ KE + + ++W TF P N + DL RD +
Sbjct: 176 NRFKEAVASKALE----------VEVKDMDWESTFFLRHLPTSNISEIPDLSQEYRDAMK 325
Query: 172 EYGEYVTKLGMTLFELLSEALGLHPNHLKDMGC-AKG--ILSLSHYYPACPEPELTMGTT 228
E+ + + KL L +LL E LGL +LK+ +KG + YPACP+PEL G
Sbjct: 326 EFAKKLEKLAEELLDLLCENLGLEKGYLKNAFYGSKGPNFGTKVANYPACPKPELVKGLR 505
Query: 229 KHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHK 288
H+D+ + LLLQDD + GLQ+ ++W+D+PP+ ++VVN+GD ++++TN ++KSVEH+
Sbjct: 506 AHTDAGGIILLLQDDKVSGLQLLKDDQWVDVPPMRHSIVVNLGDQIEVITNGRYKSVEHR 685
Query: 289 VLANIIGPRVSVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNT 344
V+A G R+SVA F+N ++ + E +++ Y + +Y+ Y T
Sbjct: 686 VVARTDGTRMSVASFYNPANDAVIYPAPALLEKEAQETEQVYPKFVFEDYMKLYAT 853
>TC209692 weakly similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (36%)
Length = 1229
Score = 157 bits (398), Expect = 5e-39
Identities = 104/348 (29%), Positives = 170/348 (47%), Gaps = 8/348 (2%)
Frame = +2
Query: 19 VKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFANFGNDPKTRHEITSKIRE 78
++ L+ K + +P + + H +P I+ + E+ K+
Sbjct: 32 IQELIKKPLTSVPQRYIQLHNNEPSLLAGETFSHALPTINLKKLIHGEDIELEL-EKLTS 208
Query: 79 ACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRDPMRPFSYNSNFNLYS 138
AC WGFFQ+V HGI V++ + + V FF +E K + +RP +
Sbjct: 209 ACRDWGFFQLVEHGISSVVMKTLEDEVEGFFMLPMEEKMKY----KVRPGDVEGYGTVIG 376
Query: 139 SS--ALNWRDTFVCSLAPNAPKPQ----DLPLVCRDELLEYGEYVTKLGMTLFELLSEAL 192
S L+W D + + + +LP R+ L Y E + L M L LL + L
Sbjct: 377 SEDQKLDWGDRLFMKINXRSIRNPHLFPELPSSLRNILELYIEELQNLAMILMGLLGKTL 556
Query: 193 GLHPNHLKDMGCAKGILSLSH-YYPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQVF 251
+ L+ GI ++ YYP CP+PEL MG + HSD+ +T+L Q + + GLQ+
Sbjct: 557 KIEKRELEVF--EDGIQNMRMTYYPPCPQPELVMGLSAHSDATGITILNQMNGVNGLQIK 730
Query: 252 HQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSVEHKVLANIIGPRVSVACFFNADLNSF 311
WI + I +ALVVN+GD++++++N +KSVEH+ N R+SVA FF L F
Sbjct: 731 KDGVWIPVNVISEALVVNIGDIIEIMSNGAYKSVEHRATVNSEKERISVAMFF---LPKF 901
Query: 312 SNQYGPIKEFLSEDNPPKYKEITISEYVA-YYNTKGIDGISALQHFRV 358
++ GP + ++PP +K I + EY+ Y+ ++G S L+H R+
Sbjct: 902 QSEIGPAVSLTNPEHPPLFKRIVVEEYIKDYFTHNKLNGKSYLEHMRI 1045
>TC205989 similar to UP|Q9FLV0 (Q9FLV0) Flavanone 3-hydroxylase-like protein,
partial (74%)
Length = 1192
Score = 156 bits (394), Expect = 2e-38
Identities = 90/263 (34%), Positives = 146/263 (55%), Gaps = 3/263 (1%)
Frame = +2
Query: 98 LEDMRNGVIRFFEQDVEVKKELYSRDPMRPFSYNSNFNLYSSSALNWRDTFVCSLAP--- 154
+E++ +G FF+ VE K +LYS D + +++FN+ + NWRD P
Sbjct: 11 MEEVAHG---FFKLPVEEKLKLYSEDTSKTMRLSTSFNVKKETVRNWRDYLRLHCYPLEK 181
Query: 155 NAPKPQDLPLVCRDELLEYGEYVTKLGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHY 214
AP+ P ++ + EY + +LG+ + E +SE+LGL +++K++ +G +Y
Sbjct: 182 YAPEWPSNPPSFKETVTEYCTIIRELGLRIQEYISESLGLEKDYIKNVLGEQGQHMAVNY 361
Query: 215 YPACPEPELTMGTTKHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLL 274
YP CPEPELT G H+D LT+LLQD + GLQV KW+ + P P+A V+N+GD L
Sbjct: 362 YPPCPEPELTYGLPGHTDPNALTILLQDLQVAGLQVLKDGKWLAVSPQPNAFVINIGDQL 541
Query: 275 QLVTNDKFKSVEHKVLANIIGPRVSVACFFNADLNSFSNQYGPIKEFLSEDNPPKYKEIT 334
Q ++N +KSV H+ + N+ PR+SVA F + + + P+ E SE Y+ T
Sbjct: 542 QALSNGLYKSVWHRAVVNVEKPRLSVASFLCPNDEALISPAKPLTEHGSE---AVYRGFT 712
Query: 335 ISEYVAYYNTKGIDGISALQHFR 357
+EY + ++ +D L+ F+
Sbjct: 713 YAEYYKKFWSRNLDQEHCLELFK 781
>TC216188 similar to UP|Q84L58 (Q84L58) 1-aminocyclopropane-1-carboxylic acid
oxidase, complete
Length = 1362
Score = 155 bits (392), Expect = 3e-38
Identities = 97/307 (31%), Positives = 159/307 (51%), Gaps = 18/307 (5%)
Frame = +3
Query: 54 IPIIDFANFGNDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDV 113
+P+IDF+ + +T+ ++I CE WGFFQ++NHGIP +LE ++
Sbjct: 147 VPVIDFSKLNGEERTK--TMAQIANGCEEWGFFQLINHGIPEELLERVK----------- 287
Query: 114 EVKKELYSRDPMRPFSYNSNFNLYSSSA---------LNWRDTFVCSLAPNAPKPQDLPL 164
+V E Y + F +++ L S S ++W D V +L + P+ P
Sbjct: 288 KVASEFYKLEREENFKNSTSVKLLSDSVEKKSSEMEHVDWED--VITLLDDNEWPEKTPG 461
Query: 165 VCRDELLEYGEYVTKLGMTLFELLSEALGLHPNHLK------DMGCAKGILSLSHYYPAC 218
R+ + EY + KL L E++ E LGL ++K D A +SHY P C
Sbjct: 462 F-RETMAEYRAELKKLAEKLMEVMDENLGLTKGYIKKALNGGDGENAFFGTKVSHY-PPC 635
Query: 219 PEPELTMGTTKHSDSCFLTLLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVT 278
P PEL G H+D+ + LL QDD +GGLQ+ + +WID+ P+P+A+V+N GD +++++
Sbjct: 636 PHPELVKGLRAHTDAGGVILLFQDDKVGGLQMLKEGQWIDVQPLPNAIVINTGDQIEVLS 815
Query: 279 NDKFKSVEHKVLANIIGPRVSVACFFNADLNSFSNQYGPIKEFLSEDN---PPKYKEITI 335
N ++KS H+VLA G R S+A F+N SF P + + +++ Y +
Sbjct: 816 NGRYKSCWHRVLATPDGNRRSIASFYNP---SFKATICPAPQLVEKEDQQVDETYPKFVF 986
Query: 336 SEYVAYY 342
+Y++ Y
Sbjct: 987 GDYMSVY 1007
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,735,135
Number of Sequences: 63676
Number of extensions: 276451
Number of successful extensions: 1492
Number of sequences better than 10.0: 217
Number of HSP's better than 10.0 without gapping: 1365
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1374
length of query: 358
length of database: 12,639,632
effective HSP length: 98
effective length of query: 260
effective length of database: 6,399,384
effective search space: 1663839840
effective search space used: 1663839840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146817.6


