

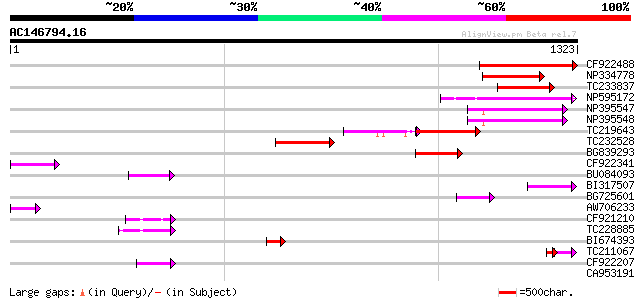
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146794.16 + phase: 0 /pseudo/partial
(1323 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922488 313 3e-85
NP334778 reverse transcriptase [Glycine max] 216 7e-56
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 204 2e-52
NP595172 polyprotein [Glycine max] 189 5e-48
NP395547 reverse transcriptase [Glycine max] 186 4e-47
NP395548 reverse transcriptase [Glycine max] 178 2e-44
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 168 2e-41
TC232528 weakly similar to UP|Q6WAY5 (Q6WAY5) Gag/pol polyprotei... 153 5e-37
BG839293 110 4e-24
CF922341 96 1e-19
BU084093 67 7e-11
BI317507 62 1e-09
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 55 2e-07
AW706233 55 3e-07
CF921210 54 5e-07
TC228885 54 5e-07
BI674393 52 2e-06
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 37 7e-06
CF922207 46 9e-05
CA953191 42 0.002
>CF922488
Length = 741
Score = 313 bits (802), Expect = 3e-85
Identities = 156/228 (68%), Positives = 180/228 (78%)
Frame = +3
Query: 1096 VPKKDGKVRMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSP 1155
V K+DGKV MCVD+RDLN AS KD FPLPHI+VLVDNT FSFMDGFSGYNQIK++P
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 1156 EDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTD 1215
ED EKT+FIT WGTFCYK M FGL N GATYQR M LF DM+HKE+EVY+DDMIVKS
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 1216 EEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMP 1275
EE+H+ L K+F RLRKY+LRLNP KC F V+S KLL FI S +GIEVD +KV+ I EM
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 1276 APQTQKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDEC 1323
P T+KQV+GFLGRLNYI RFIS + ATC P+F LL KNQ + W+ +C
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC 686
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 216 bits (549), Expect = 7e-56
Identities = 100/143 (69%), Positives = 116/143 (80%)
Frame = +3
Query: 1104 RMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSF 1163
RMCVD+RDLN+AS KDNFPLPHID+L+ N A +FSFMDGFSGYNQIKM+PED EKT+F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 1164 ITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYL 1223
IT WGTFCYKVM FGL N GATY R M LF DM+HKE+E YVD+MI KS EE+H+ L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 1224 TKMFERLRKYKLRLNPNKCTFGV 1246
+F +LRKY+LRLNP KC FG+
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 204 bits (520), Expect = 2e-52
Identities = 96/133 (72%), Positives = 113/133 (84%)
Frame = +2
Query: 1139 FSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMI 1198
FSFMDGFSGYNQI M+ ED EKT+F+T WGTF Y+VM FGL N GATYQR M LFHDM+
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 1199 HKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQ 1258
HKE+EVYVDDMI KS E +H+ L K+F RL+KY+L+LNP KCTFGV+SGKLLGFIVSQ
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 1259 KGIEVDPDKVRAI 1271
KGIE+DP+KV+A+
Sbjct: 362 KGIEIDPEKVKAL 400
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 189 bits (481), Expect = 5e-48
Identities = 112/317 (35%), Positives = 174/317 (54%)
Frame = +1
Query: 1006 LEEGVKKKIIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMA 1065
L + ++ LL Y +FA P + +H IP K PV+ + R
Sbjct: 1660 LPTNIDPELAILLHTYAQVFAVPASLPPQREQ---DHAIPLKQGSGPVKVRPYRYPHTQK 1830
Query: 1066 LKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLPH 1125
+I+ +Q+ + G + P + I+ V KKDG R C D+R LN ++KD+FP+P
Sbjct: 1831 DQIEKMIQEMLVQGIIQPSNSP-FSLPILLVKKKDGSWRFCTDYRALNAITVKDSFPMPT 2007
Query: 1126 IDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGAT 1185
+D L+D ++ FS +D SGY+QI + PEDREKT+F T G + + VMPFGL NA AT
Sbjct: 2008 VDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPAT 2187
Query: 1186 YQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFG 1245
+Q M +F + K V V+ DD+++ S + H+++L + + L++++L +KC+FG
Sbjct: 2188 FQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLSKCSFG 2367
Query: 1246 VRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISHMTATCG 1305
LG VS G+ ++ KV+A+ + P P KQ+RGFLG Y RFI G
Sbjct: 2368 DTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFLGLTGYYRRFIKSYANIAG 2547
Query: 1306 PIFKLLRKNQPIVWNDE 1322
P+ LL+K+ +WN+E
Sbjct: 2548 PLTDLLQKDS-FLWNNE 2595
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 186 bits (473), Expect = 4e-47
Identities = 95/252 (37%), Positives = 143/252 (56%), Gaps = 18/252 (7%)
Frame = +1
Query: 1068 IKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV------------------RMCVDF 1109
++ EV K ++AG + + WV+ + VPKK G RMC+D+
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 1110 RDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGT 1169
R LN+A+ KD++PLP +D ++ A+ + F+DG+SGYNQI + P+D+EKT+F P+
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 1170 FCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFER 1229
F Y+ MPFGL NA T+QR M +F DM+ K +EV++DD + L K+ +R
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 1230 LRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGR 1289
K L LN KC F V+ G +LG +S++GIEV +K+ I ++P P K + FLG
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 1290 LNYISRFISHMT 1301
+ + RFI T
Sbjct: 721 VGFYRRFIKDFT 756
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 178 bits (451), Expect = 2e-44
Identities = 90/252 (35%), Positives = 142/252 (55%), Gaps = 18/252 (7%)
Frame = +1
Query: 1068 IKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV------------------RMCVDF 1109
++ EV K ++ G + + WV+ ++ V KK+G ++C+D+
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 1110 RDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGT 1169
R LN+A+ KD+FPLP +D +++ A + F+D + GYNQI + P+D+EK +F P+G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 1170 FCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQHVEYLTKMFER 1229
F Y+ +PFGL NA T+Q M +F D++ K +EV++DD V E ++ L + +R
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 1230 LRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTQKQVRGFLGR 1289
+ L LN KC F VR G +LG +S +GIEVD K+ I ++P P K +R FLG+
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 1290 LNYISRFISHMT 1301
+ RFI T
Sbjct: 721 ARFYRRFIKDFT 756
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein (Fragment),
partial (8%)
Length = 1320
Score = 168 bits (425), Expect = 2e-41
Identities = 78/152 (51%), Positives = 109/152 (71%), Gaps = 2/152 (1%)
Frame = +3
Query: 948 NFEFPVYEAEDEEGDDI--PYEITRLLEQEKKAIQPH*EEIELINIGTEENKREIKIGAA 1005
+FE + + EDE +D+ P E+ R++ E + + PH EE EL+++G+ KRE+KIG
Sbjct: 864 DFEQKMNQTEDEGNEDVGLPPELERMVAHEDQEMGPHQEETELVDLGSGSGKREVKIGTG 1043
Query: 1006 LEEGVKKKIIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMA 1065
+ +++++I LLR+Y DIFAWSY+DMPGL IV+HR+P PEC PV+QKLRR P+ +
Sbjct: 1044 ITAPIREELIILLRDYQDIFAWSYQDMPGLSSDIVQHRLPLNPECSPVKQKLRRMKPETS 1223
Query: 1066 LKIKSEVQKQIDAGFLMTVEYPEWVANIVPVP 1097
LKIK EV+K DAGFL YP+WVANIVP+P
Sbjct: 1224 LKIKEEVKK*FDAGFLAVARYPKWVANIVPIP 1319
Score = 66.6 bits (161), Expect = 7e-11
Identities = 66/212 (31%), Positives = 87/212 (40%), Gaps = 29/212 (13%)
Frame = +1
Query: 779 PWIHDAGAVTSTLHQKLKFAKSGKLVTIHGEEAYLVSQLSSFSCIEAGSAE-GTAFQGLT 837
PWIH G V STLHQKLKF G LV + GEE LVS SS +EA TAFQ
Sbjct: 1 PWIHSVGVVPSTLHQKLKFVVEGHLVIVSGEEDILVSCPSSMPYVEAAEESLETAFQSFE 180
Query: 838 VEGTEPKRDGTAMASLKD-----AQRAVQEGQAAGWG---------RLIQLRENKHKEGL 883
V L D A+ + G G G LI + N+ K GL
Sbjct: 181 VVSISSVDSLFGQPCLSDAAVMMARVMLGNGYEPGMGLGKDNGGITSLINTQGNRGKYGL 360
Query: 884 GFSPTSG---VSTGAFYSAGFVNAITEEATGFGPRPV---FVIPG--------GIARDWD 929
G+ PT S ++G + +E+ G P + F+ G I D
Sbjct: 361 GYKPTQADMKRSIAGRKNSGQSSRWRQESEGSPPCHISRSFISAGLGDKGQVFAICED-- 534
Query: 930 AIDIPSIMHVSDNPAFPPNFEFPVYEAEDEEG 961
D+PS + + P PP+F+ + E+ G
Sbjct: 535 --DVPSTLDLV-RPC-PPDFQLGNWRVEERSG 618
>TC232528 weakly similar to UP|Q6WAY5 (Q6WAY5) Gag/pol polyprotein
(Fragment), partial (3%)
Length = 449
Score = 153 bits (386), Expect = 5e-37
Identities = 72/139 (51%), Positives = 104/139 (74%)
Frame = +1
Query: 620 SKISILSLLLSSAAHRDTLLKVLEQAYVDHEVTVDHFGSIVGNITACSNLWFSEDELPEA 679
+++S+L LL+SS HR L+KVL +A+V +++V+ FG +V NITA + L F+E+E+P
Sbjct: 31 ARVSLLELLMSSEPHRALLVKVLNEAHVAQDISVEGFGGLVNNITANNYLAFAEEEIPAE 210
Query: 680 GKHHNLALHISVNCKSDMISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFLVKAFDG 739
G+ HN ALH+SV C +++ VL+D G SLNVMPKSTLD+L + + L+ S+ +V+AFDG
Sbjct: 211 GRGHNKALHVSVKCMDHIVAKVLIDNGYSLNVMPKSTLDKLPFNASHLKPSSMVVRAFDG 390
Query: 740 TRKSVLGEIDLPITIGPET 758
TR+ V GEIDLP+ IGP T
Sbjct: 391 TRREVRGEIDLPVQIGPHT 447
>BG839293
Length = 781
Score = 110 bits (275), Expect = 4e-24
Identities = 50/110 (45%), Positives = 75/110 (67%), Gaps = 2/110 (1%)
Frame = +1
Query: 948 NFEFPVYEAEDEEGDDI--PYEITRLLEQEKKAIQPH*EEIELINIGTEENKREIKIGAA 1005
NFE + EDE +D+ P E+ R++ E + + PH EE EL+++G KRE+KIG
Sbjct: 400 NFEQETSQTEDEGNEDVGLPPELERMVAHEDQEMGPHQEETELVDLGIGSGKREVKIGTG 579
Query: 1006 LEEGVKKKIIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQ 1055
+ +++++I LL++Y DIFAWSY+DMPGL IV+H++P PEC PV+Q
Sbjct: 580 ITAPIREELIILLKDYQDIFAWSYQDMPGLSSDIVQHQLPLNPECSPVKQ 729
>CF922341
Length = 675
Score = 95.5 bits (236), Expect = 1e-19
Identities = 48/119 (40%), Positives = 70/119 (58%), Gaps = 4/119 (3%)
Frame = +1
Query: 1 MEELAK--ELRHEIKANRGNADSF--KTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNH 56
M E+ K L ++A G D ++L LV + P KFK+ DFD+Y G TCP+NH
Sbjct: 289 MAEMGKLDHLEEGLRAIEGGEDYAFANLEELFLVPNIITPPKFKVLDFDKYKGTTCPKNH 468
Query: 57 IIKYVRKMGNYKDNDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAVAFKSHYGFN 115
+ Y +KMG Y ++ L+IH FQ+SL A WYT+L + +H++ +L VAF Y +N
Sbjct: 469 LKMYCQKMGAYAKDEELLIHSFQESLTGVAVTWYTNLEPSRVHSWKDLMVAFVRQYQYN 645
>BU084093
Length = 421
Score = 66.6 bits (161), Expect = 7e-11
Identities = 40/115 (34%), Positives = 53/115 (45%), Gaps = 7/115 (6%)
Frame = -1
Query: 277 PGQPQVPVNAIAQQM---KQQLPVQQQQQNQQARPT----FPPIPMLYAELLPTLLHRGH 329
P QP+ P A Q+ PV + P F P+PM Y +LLP+L+
Sbjct: 346 PVQPKAPAQREAPQVPTPNTTRPVGNSNTTRNFPPRPFQEFTPLPMTYEDLLPSLIANHL 167
Query: 330 CTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTF 384
G+ P P + + C +H G GH VE C ALKY V+ L+D G LTF
Sbjct: 166 AVVTPGRVLQPPFPKWYDPNATCKYHGGVPGHSVEKCLALKYKVQHLMDAGWLTF 2
>BI317507
Length = 359
Score = 62.4 bits (150), Expect = 1e-09
Identities = 33/114 (28%), Positives = 58/114 (49%)
Frame = -1
Query: 1208 DMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDK 1267
++++ S D + H+ +LT + + L+K +L N KC F + + LG ++S+ + +D +K
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 1268 VRAIREMPAPQTQKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPIVWND 1321
V+++ E P P+ K+V FL Y +FI L KN W D
Sbjct: 173 VKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLTKNDGFKWGD 12
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 55.5 bits (132), Expect = 2e-07
Identities = 30/90 (33%), Positives = 48/90 (53%)
Frame = -3
Query: 1042 HRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDG 1101
H++ + V Q+ R+ + + EV K A F+ + Y + ++V V K +G
Sbjct: 277 HKLAICNDVKLVTQRKRKIREERCQTV*QEVVKLAIASFIRDINYST*LFSVVMVKKPNG 98
Query: 1102 KVRMCVDFRDLNKASLKDNFPLPHIDVLVD 1131
K R+C D+ DLN A KD +PLP+ID + D
Sbjct: 97 KWRICTDYIDLN*ACPKDAYPLPNIDHMTD 8
>AW706233
Length = 376
Score = 54.7 bits (130), Expect = 3e-07
Identities = 27/71 (38%), Positives = 39/71 (54%), Gaps = 2/71 (2%)
Frame = -3
Query: 3 ELAKELRHEIKANRGNADSF--KTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKY 60
E L+ KA G D ++L LV + P KFK+ +FD+Y G TCP+NH+ Y
Sbjct: 347 EKLDHLKERFKAIEGGQDYAFANLEELFLVXNIISPPKFKVLNFDKYKGTTCPKNHLKMY 168
Query: 61 VRKMGNYKDND 71
+KMG Y ++
Sbjct: 167 CQKMGAYAKDE 135
>CF921210
Length = 790
Score = 53.9 bits (128), Expect = 5e-07
Identities = 41/118 (34%), Positives = 55/118 (45%)
Frame = +2
Query: 270 YHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQQQNQQARPTFPPIPMLYAELLPTLLHRGH 329
YH+ LP Q + P A + Q++ ++ + F PIP+ YA+LL LL
Sbjct: 236 YHKVCLPHFQ*RTPPLAQTKNTNQEVNFAARKPVE-----FTPIPVSYADLLSYLLDNSM 400
Query: 330 CTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTFENN 387
K PL + S+ C GAL H +E C ALK V+ LID G L FE N
Sbjct: 401 VAITLAKVHQPPLF*GYDSNATCG---GALRHSIEHCRALKRKVQGLIDAGWLKFEEN 565
>TC228885
Length = 901
Score = 53.9 bits (128), Expect = 5e-07
Identities = 42/135 (31%), Positives = 58/135 (42%)
Frame = -3
Query: 253 PSYPYVPYSQHPFFPPFYHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQQQNQQARPTFPP 312
P +P + H P H+ LP Q + P A + Q++ ++ + F P
Sbjct: 761 PHHPVI----HKGHP*INHKVCLPHFQ*RTPPLAQTKNTNQEMNFAARKPVE-----FTP 609
Query: 313 IPMLYAELLPTLLHRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYI 372
IP+ YA+LLP LL K P + S+ C H A G +E ALK
Sbjct: 608 IPVSYADLLPYLLDNSMVAITLAKVHQPPFLREYDSNAMCACHGEAPGRSIEHYRALKRK 429
Query: 373 VKKLIDQGKLTFENN 387
V+ LID G L FE N
Sbjct: 428 VQGLIDAGWLKFEEN 384
>BI674393
Length = 152
Score = 52.0 bits (123), Expect = 2e-06
Identities = 22/44 (50%), Positives = 37/44 (84%)
Frame = +1
Query: 599 EILRLIKRSDYKIVDQLLQTPSKISILSLLLSSAAHRDTLLKVL 642
E LR+I++S++K+++QL +TP+++S+L LL+SS HR L+KVL
Sbjct: 16 EFLRIIQQSEFKVIEQLNKTPARVSLLELLMSSEPHRALLVKVL 147
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 36.6 bits (83), Expect(2) = 7e-06
Identities = 12/25 (48%), Positives = 21/25 (84%)
Frame = +2
Query: 1253 GFIVSQKGIEVDPDKVRAIREMPAP 1277
GF+V + G+++DP+K++AI+E P P
Sbjct: 29 GFVVGRNGVQMDPEKIKAIQEWPPP 103
Score = 32.7 bits (73), Expect(2) = 7e-06
Identities = 12/51 (23%), Positives = 26/51 (50%)
Frame = +1
Query: 1272 REMPAPQTQKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDE 1322
R P ++ +R F G ++ RF+ + + P+ +L++KN W ++
Sbjct: 88 RMAPTLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEK 240
>CF922207
Length = 616
Score = 46.2 bits (108), Expect = 9e-05
Identities = 31/95 (32%), Positives = 40/95 (41%), Gaps = 3/95 (3%)
Frame = -1
Query: 296 PVQQQQQN---QQARPTFPPIPMLYAELLPTLLHRGHCTTRQGKPPPDPLPPRFRSDLKC 352
P + QQ+ Q+A P L+P L K P P + S+ C
Sbjct: 613 PKKHQQKGILQQKACRVHPKFRCHMLNLIPYPLDNSMVAITPTKVPQPPFFREYDSNATC 434
Query: 353 DFHQGALGHDVEGCYALKYIVKKLIDQGKLTFENN 387
+H GA GH +E C K+ V LID G L FE N
Sbjct: 433 AYHGGAPGHSIEHCMTPKHKV*SLIDTG*LKFEEN 329
>CA953191
Length = 422
Score = 42.0 bits (97), Expect = 0.002
Identities = 28/58 (48%), Positives = 32/58 (54%), Gaps = 1/58 (1%)
Frame = -3
Query: 768 INASYSCLLGRPW-IHDAGAVTSTLHQKLKFAKSGKLVTIHGEEAYLVSQLSSFSCIE 824
I +Y+ L GRPW IH V STLH K K GKLV I +E LV + SS IE
Sbjct: 420 ITPTYNGLQGRPWRIHCVKLVPSTLH*K*KIVIDGKLVIIFVKEDLLVGEPSSTPYIE 247
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 57,112,566
Number of Sequences: 63676
Number of extensions: 843999
Number of successful extensions: 6261
Number of sequences better than 10.0: 144
Number of HSP's better than 10.0 without gapping: 5533
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6024
length of query: 1323
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1214
effective length of database: 5,698,948
effective search space: 6918522872
effective search space used: 6918522872
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146794.16


