

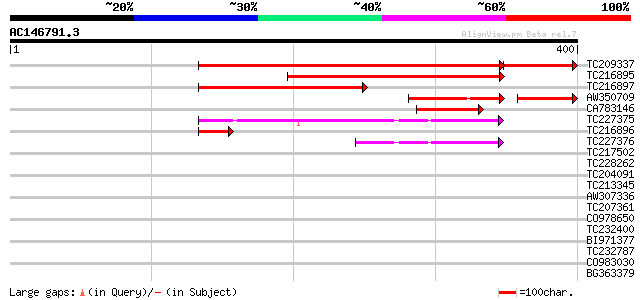
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146791.3 - phase: 0 /pseudo
(400 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209337 homologue to UP|KPR2_ARATH (Q42583) Ribose-phosphate py... 417 e-117
TC216895 similar to UP|Q8L7T8 (Q8L7T8) At2g44530/F16B22.2, parti... 258 4e-69
TC216897 homologue to UP|Q9XG98 (Q9XG98) Phosphoribosyl pyrophos... 225 3e-59
AW350709 similar to PIR|S71262|S712 ribose-phosphate diphosphoki... 85 8e-23
CA783146 homologue to GP|4902849|emb| phosphoribosyl pyrophospha... 67 1e-11
TC227375 similar to PIR|T52589|T52589 ribose-phosphate diphospho... 67 2e-11
TC216896 homologue to UP|Q9XG98 (Q9XG98) Phosphoribosyl pyrophos... 53 2e-07
TC227376 similar to UP|Q9XGA1 (Q9XGA1) Phosphoribosyl pyrophosph... 47 2e-05
TC217502 similar to GB|CAB96651.1|8953378|ATF2I11 adenine phosph... 37 0.012
TC228262 similar to UP|Q9MAL4 (Q9MAL4) F27F5.2, partial (4%) 36 0.027
TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1... 35 0.060
TC213345 similar to GB|AAP13819.1|37955428|AY254320 DVL4 {Arabid... 35 0.078
AW307336 weakly similar to GP|22202765|dbj P0039H02.8 {Oryza sat... 35 0.078
TC207361 similar to UP|Q9FMU9 (Q9FMU9) Similarity to ATFP3, part... 35 0.078
CO978650 34 0.10
TC232400 weakly similar to UP|Q8W2K5 (Q8W2K5) Phragmoplastin-int... 33 0.17
BI971377 33 0.17
TC232787 UP|O48239 (O48239) Cytochrome b (Fragment), partial (8%) 33 0.17
CO983030 32 0.51
BG363379 similar to GP|2224419|dbj|B ORF67 {Chlorella vulgaris},... 31 1.1
>TC209337 homologue to UP|KPR2_ARATH (Q42583) Ribose-phosphate
pyrophosphokinase 2 (Phosphoribosyl pyrophosphate
synthetase 2) (PRS II) , partial (72%)
Length = 942
Score = 417 bits (1072), Expect = e-117
Identities = 208/216 (96%), Positives = 212/216 (97%)
Frame = +1
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGCNVYLIQPTCPPANENLMEL++MIDACRRASAKNITAVIPYFGYARADR
Sbjct: 55 IYVQLQESVRGCNVYLIQPTCPPANENLMELKIMIDACRRASAKNITAVIPYFGYARADR 234
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 253
KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT
Sbjct: 235 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKT 414
Query: 254 ISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVM 313
ISS+DLVVVSPDVGGVARARAFAKKLSDAPLA DKRR GHNVAEVMNLIGDVKGKVAVM
Sbjct: 415 ISSSDLVVVSPDVGGVARARAFAKKLSDAPLAXXDKRRHGHNVAEVMNLIGDVKGKVAVM 594
Query: 314 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
VDDMIDTAGTIAKGAALLHEEGAREVYACCTH VFS
Sbjct: 595 VDDMIDTAGTIAKGAALLHEEGAREVYACCTHAVFS 702
Score = 57.4 bits (137), Expect = 1e-08
Identities = 29/52 (55%), Positives = 35/52 (66%)
Frame = +3
Query: 349 SLIMFVLLYLQPSCNREAVQRLISRGDNHEHYSGSREELFPPVNYSYRSKPV 400
S+ M Q S N E V+ L+S GD H+H+S REELFPPVNYSY SKP+
Sbjct: 669 SVCMLHSCRFQSSRN*EVVRWLVS*GDYHKHHSSCREELFPPVNYSYCSKPI 824
>TC216895 similar to UP|Q8L7T8 (Q8L7T8) At2g44530/F16B22.2, partial (55%)
Length = 1024
Score = 258 bits (658), Expect = 4e-69
Identities = 132/153 (86%), Positives = 139/153 (90%)
Frame = +2
Query: 197 GRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASKTISS 256
GRES+ LVAN +AGA+RVLACDLHS SMGY DIPVDHV+ QPVILDYLASKTI S
Sbjct: 2 GRESMLXXLVANXXXEAGANRVLACDLHSXXSMGYXDIPVDHVYGQPVILDYLASKTICS 181
Query: 257 NDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGDVKGKVAVMVDD 316
+DLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRR GHNVAEVMNLIGDV+GKVAVMVDD
Sbjct: 182 DDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRHGHNVAEVMNLIGDVRGKVAVMVDD 361
Query: 317 MIDTAGTIAKGAALLHEEGAREVYACCTHGVFS 349
MIDTAGTIAKGAALLH+EGAREVYAC TH VFS
Sbjct: 362 MIDTAGTIAKGAALLHQEGAREVYACTTHAVFS 460
>TC216897 homologue to UP|Q9XG98 (Q9XG98) Phosphoribosyl pyrophosphate
synthase , partial (48%)
Length = 738
Score = 225 bits (573), Expect = 3e-59
Identities = 109/119 (91%), Positives = 117/119 (97%)
Frame = +2
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+YVQLQESVRGC+V+L+QPTCPPANENLMEL +MIDACRRASAKNITAVIPYFGYARADR
Sbjct: 380 IYVQLQESVRGCDVFLVQPTCPPANENLMELLIMIDACRRASAKNITAVIPYFGYARADR 559
Query: 194 KTQGRESIAAKLVANLITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPVILDYLASK 252
KTQGRESIAAKLVANLIT+AGA+RVLACDLHSGQSMGYFDIPVDHV+ QPVILDYLASK
Sbjct: 560 KTQGRESIAAKLVANLITEAGANRVLACDLHSGQSMGYFDIPVDHVYGQPVILDYLASK 736
>AW350709 similar to PIR|S71262|S712 ribose-phosphate diphosphokinase (EC
2.7.6.1) isoform II [validated] - Arabidopsis thaliana,
partial (28%)
Length = 376
Score = 85.1 bits (209), Expect(2) = 8e-23
Identities = 48/68 (70%), Positives = 52/68 (75%)
Frame = -3
Query: 282 APLAIVDKRRSGHNVAEVMNLIGDVKGKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYA 341
APLAIVDKR NVAEVMNLIGDVK KVAVMVDDMID+AGT ++ EGAR+VYA
Sbjct: 374 APLAIVDKRHHXXNVAEVMNLIGDVKEKVAVMVDDMIDSAGT-SERVQHXX*EGARKVYA 198
Query: 342 CCTHGVFS 349
C H VFS
Sbjct: 197 *CNHTVFS 174
Score = 40.0 bits (92), Expect(2) = 8e-23
Identities = 18/42 (42%), Positives = 28/42 (65%)
Frame = -2
Query: 359 QPSCNREAVQRLISRGDNHEHYSGSREELFPPVNYSYRSKPV 400
Q S N + V+ L+S G+ H+H+S + + FP +NYSY SK +
Sbjct: 177 QSSSN*KVVRWLVS*GNYHKHHSRAEKNYFPQLNYSYSSKHI 52
>CA783146 homologue to GP|4902849|emb| phosphoribosyl pyrophosphate synthase
{Spinacia oleracea}, partial (10%)
Length = 444
Score = 67.4 bits (163), Expect = 1e-11
Identities = 36/47 (76%), Positives = 38/47 (80%)
Frame = +1
Query: 288 DKRRSGHNVAEVMNLIGDVKGKVAVMVDDMIDTAGTIAKGAALLHEE 334
DKRR HNVAEVMNLIGDVKGKVAVMVDDMIDTAG + + L EE
Sbjct: 1 DKRRPLHNVAEVMNLIGDVKGKVAVMVDDMIDTAGMPSSLSNLNFEE 141
>TC227375 similar to PIR|T52589|T52589 ribose-phosphate diphosphokinase
[imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (93%)
Length = 1334
Score = 66.6 bits (161), Expect = 2e-11
Identities = 61/222 (27%), Positives = 109/222 (48%), Gaps = 7/222 (3%)
Frame = +1
Query: 134 VYVQLQESVRGCNVYLIQPTCPPANENLMELQVMIDACRRASAKNITAVIPYFGYARADR 193
+Y+ E +RG +V + PA+ + E +I A R + T V+P+F +R
Sbjct: 310 IYINNAEELRGQHVAFLASFSSPAH--VFEQLSVIYALPRLFVASFTLVLPFFPTGSFER 483
Query: 194 -KTQGRESIA---AKLVANL-ITKAGADRVLACDLHSGQSMGYFDIPVDHVHCQPV-ILD 247
+ +G + A A++++N+ I++ G ++ D+H+ Q YF V + + +L
Sbjct: 484 MEEEGDVATAFTLARMLSNIPISRGGPTSLVIYDIHALQERFYFGDEVLPLFETGIPLLK 663
Query: 248 YLASKTISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLI-GDV 306
S+ ++++V+ PD G R F K + L + K R G ++ L G+V
Sbjct: 664 QRLSQLPDADNVVIAFPDDGAWKR---FHKLFDNFSLVVCTKVREGDK--RIVRLKEGNV 828
Query: 307 KGKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVF 348
G V+VDD++ + GT+ + +L GA +V A THGVF
Sbjct: 829 SGHHVVIVDDLVQSGGTLIECQKVLAANGAAKVSAYVTHGVF 954
>TC216896 homologue to UP|Q9XG98 (Q9XG98) Phosphoribosyl pyrophosphate
synthase , partial (20%)
Length = 448
Score = 53.1 bits (126), Expect = 2e-07
Identities = 21/25 (84%), Positives = 25/25 (100%)
Frame = +3
Query: 134 VYVQLQESVRGCNVYLIQPTCPPAN 158
+YVQLQESVRGC+V+L+QPTCPPAN
Sbjct: 372 IYVQLQESVRGCDVFLVQPTCPPAN 446
>TC227376 similar to UP|Q9XGA1 (Q9XGA1) Phosphoribosyl pyrophosphate synthase
isozyme 4 , partial (49%)
Length = 797
Score = 46.6 bits (109), Expect = 2e-05
Identities = 33/105 (31%), Positives = 53/105 (50%), Gaps = 1/105 (0%)
Frame = +2
Query: 245 ILDYLASKTISSNDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLI- 303
+L S+ ++++V+ PD G R F K+ L + K R G ++ L
Sbjct: 2 LLKQRLSQLPDADNVVIAFPDDGAWKR---FHKQFDHFSLXVCTKVREGDK--RIVRLKE 166
Query: 304 GDVKGKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYACCTHGVF 348
G+V G V+VDD++ + GT+ + +L GA +V A THGVF
Sbjct: 167 GNVSGHHVVIVDDLVQSGGTLIECQKVLAANGAAKVSAYVTHGVF 301
>TC217502 similar to GB|CAB96651.1|8953378|ATF2I11 adenine
phosphoribosyltransferase-like protein {Arabidopsis
thaliana;} , partial (96%)
Length = 1068
Score = 37.4 bits (85), Expect = 0.012
Identities = 16/40 (40%), Positives = 24/40 (60%)
Frame = +1
Query: 308 GKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYACCTHGV 347
G+ A+++DD++ T GT++ G LL GA V C GV
Sbjct: 619 GERAIIIDDLVATGGTLSAGVKLLERVGAEVVECACVIGV 738
>TC228262 similar to UP|Q9MAL4 (Q9MAL4) F27F5.2, partial (4%)
Length = 657
Score = 36.2 bits (82), Expect = 0.027
Identities = 29/87 (33%), Positives = 41/87 (46%), Gaps = 11/87 (12%)
Frame = -2
Query: 5 LFQLQHTSSSTMPFAPSSFF-----------SGVSHGLTFPTLGFVDLSRSRTVAYNTVV 53
LF++ ++ MPF FF VSH L T+ F+ L + + T
Sbjct: 296 LFRIHIIYTALMPFPMFPFFILFFFLLLFVTMAVSHVLII-TICFILLITFLRLIFFTFS 120
Query: 54 ITFFLFFILLCFIFVFFFTFFIFLLEM 80
I FF FF+LL F+F FF F +FL +
Sbjct: 119 ILFF-FFLLLPFLFSFFLLFSLFLFTL 42
>TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(70%)
Length = 2075
Score = 35.0 bits (79), Expect = 0.060
Identities = 14/28 (50%), Positives = 20/28 (71%)
Frame = -2
Query: 49 YNTVVITFFLFFILLCFIFVFFFTFFIF 76
++ VV FF FF+LL F+ + FF FF+F
Sbjct: 259 FDDVVFFFFFFFLLLFFLVLLFFFFFLF 176
Score = 30.0 bits (66), Expect = 1.9
Identities = 13/30 (43%), Positives = 20/30 (66%), Gaps = 4/30 (13%)
Frame = -2
Query: 52 VVITFFLFFILLCFIFVFF----FTFFIFL 77
+V+ FF FF+ F+++FF F FF+FL
Sbjct: 208 LVLLFFFFFLFDLFLYLFFLNFLFDFFVFL 119
>TC213345 similar to GB|AAP13819.1|37955428|AY254320 DVL4 {Arabidopsis
thaliana;} , partial (93%)
Length = 426
Score = 34.7 bits (78), Expect = 0.078
Identities = 12/25 (48%), Positives = 20/25 (80%)
Frame = +2
Query: 56 FFLFFILLCFIFVFFFTFFIFLLEM 80
FFL+F L CF ++F F+FF+ L+++
Sbjct: 278 FFLYFFLSCFFYLFLFSFFLPLIQI 352
>AW307336 weakly similar to GP|22202765|dbj P0039H02.8 {Oryza sativa
(japonica cultivar-group)}, partial (18%)
Length = 371
Score = 34.7 bits (78), Expect = 0.078
Identities = 20/50 (40%), Positives = 23/50 (46%)
Frame = -3
Query: 27 VSHGLTFPTLGFVDLSRSRTVAYNTVVITFFLFFILLCFIFVFFFTFFIF 76
VSH F L F + + N V FF FF+ F F FFF FF F
Sbjct: 228 VSHFTLFGCLPFFAIVKKT----NFEVFFFFFFFLFFFFFFFFFFFFFFF 91
Score = 32.0 bits (71), Expect = 0.51
Identities = 13/21 (61%), Positives = 13/21 (61%)
Frame = -3
Query: 56 FFLFFILLCFIFVFFFTFFIF 76
FFLFF F F FFF FF F
Sbjct: 144 FFLFFFFFFFFFFFFFFFFFF 82
Score = 30.8 bits (68), Expect = 1.1
Identities = 12/22 (54%), Positives = 13/22 (58%)
Frame = -1
Query: 55 TFFLFFILLCFIFVFFFTFFIF 76
+FF FF F F FFF FF F
Sbjct: 140 SFFFFFFFFFFFFFFFFFFFFF 75
Score = 28.9 bits (63), Expect = 4.3
Identities = 11/20 (55%), Positives = 12/20 (60%)
Frame = -2
Query: 56 FFLFFILLCFIFVFFFTFFI 75
FF FF F F FFF FF+
Sbjct: 130 FFFFFFFFFFFFFFFFFFFL 71
>TC207361 similar to UP|Q9FMU9 (Q9FMU9) Similarity to ATFP3, partial (53%)
Length = 1127
Score = 34.7 bits (78), Expect = 0.078
Identities = 25/70 (35%), Positives = 33/70 (46%), Gaps = 1/70 (1%)
Frame = -3
Query: 4 SLFQLQHTSSSTMPFAPSSFFSGVSHGLTFPTLGFVDLSRSRTVAYNTVVITFFLFFIL- 62
+LFQLQH FF G F F+ + S T +++TFFL F+L
Sbjct: 852 TLFQLQHCG----------FFFGFLFCFGFTLSLFLLVFFSLTTPLLILLLTFFLCFLLF 703
Query: 63 LCFIFVFFFT 72
CF F FFF+
Sbjct: 702 FCFFFFFFFS 673
>CO978650
Length = 709
Score = 34.3 bits (77), Expect = 0.10
Identities = 16/38 (42%), Positives = 19/38 (49%)
Frame = -2
Query: 39 VDLSRSRTVAYNTVVITFFLFFILLCFIFVFFFTFFIF 76
V++ S Y +TFF FF F F FFF FF F
Sbjct: 141 VEVH*SFXXXYEKWYMTFFFFFFFFFFFFFFFFFFFFF 28
Score = 33.5 bits (75), Expect = 0.17
Identities = 14/23 (60%), Positives = 14/23 (60%)
Frame = -1
Query: 56 FFLFFILLCFIFVFFFTFFIFLL 78
FF FF F F FFF FF FLL
Sbjct: 70 FFFFFFFFFFFFFFFFFFFFFLL 2
Score = 28.5 bits (62), Expect = 5.6
Identities = 14/36 (38%), Positives = 17/36 (46%)
Frame = -1
Query: 41 LSRSRTVAYNTVVITFFLFFILLCFIFVFFFTFFIF 76
LSRS + + + FF F F FFF FF F
Sbjct: 145 LSRSALIFXXXI*KMVYDFFFFFFFFFFFFFFFFFF 38
>TC232400 weakly similar to UP|Q8W2K5 (Q8W2K5) Phragmoplastin-interacting
protein PHIP1, partial (8%)
Length = 621
Score = 33.5 bits (75), Expect = 0.17
Identities = 13/27 (48%), Positives = 18/27 (66%)
Frame = -1
Query: 48 AYNTVVITFFLFFILLCFIFVFFFTFF 74
++++ V+ FFLF L F F FFF FF
Sbjct: 387 SFSSAVVDFFLFLFLFFFCFCFFFLFF 307
>BI971377
Length = 418
Score = 33.5 bits (75), Expect = 0.17
Identities = 15/35 (42%), Positives = 22/35 (62%)
Frame = -2
Query: 53 VITFFLFFILLCFIFVFFFTFFIFLLEM*YVGSIK 87
V+ F+ F L FIF+FFF FF F +* + S++
Sbjct: 132 VLLFYFFXXLFFFIFLFFFFFFFFF*NV*SLASMR 28
>TC232787 UP|O48239 (O48239) Cytochrome b (Fragment), partial (8%)
Length = 608
Score = 33.5 bits (75), Expect = 0.17
Identities = 14/23 (60%), Positives = 14/23 (60%)
Frame = -3
Query: 56 FFLFFILLCFIFVFFFTFFIFLL 78
FF FF F F FFF FF FLL
Sbjct: 423 FFFFFFFFFFFFFFFFFFFFFLL 355
Score = 30.4 bits (67), Expect = 1.5
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = -2
Query: 56 FFLFFILLCFIFVFFFTFFIFLL 78
FF FF F F FFF FFI L+
Sbjct: 415 FFFFFFFFFFFFFFFFFFFIELV 347
Score = 29.6 bits (65), Expect = 2.5
Identities = 12/23 (52%), Positives = 13/23 (56%)
Frame = -3
Query: 53 VITFFLFFILLCFIFVFFFTFFI 75
V FF FF F F FFF FF+
Sbjct: 426 VFFFFFFFFFFFFFFFFFFFFFL 358
>CO983030
Length = 505
Score = 32.0 bits (71), Expect = 0.51
Identities = 13/21 (61%), Positives = 13/21 (61%)
Frame = -1
Query: 56 FFLFFILLCFIFVFFFTFFIF 76
FFLFF F F FFF FF F
Sbjct: 295 FFLFFFFFFFFFFFFFFFFFF 233
Score = 29.6 bits (65), Expect = 2.5
Identities = 13/26 (50%), Positives = 15/26 (57%), Gaps = 5/26 (19%)
Frame = -2
Query: 56 FFLFFI-----LLCFIFVFFFTFFIF 76
FF+FF+ CF F FFF FF F
Sbjct: 327 FFVFFLFXXFFFFCFFFFFFFFFFFF 250
Score = 29.3 bits (64), Expect = 3.3
Identities = 11/20 (55%), Positives = 12/20 (60%)
Frame = -3
Query: 57 FLFFILLCFIFVFFFTFFIF 76
F FF+ F F FFF FF F
Sbjct: 299 FFFFVFFFFFFFFFFFFFFF 240
Score = 29.3 bits (64), Expect = 3.3
Identities = 11/24 (45%), Positives = 12/24 (49%)
Frame = -1
Query: 53 VITFFLFFILLCFIFVFFFTFFIF 76
+ FF F F F FFF FF F
Sbjct: 310 IFXFFFFLFFFFFFFFFFFFFFFF 239
Score = 28.5 bits (62), Expect = 5.6
Identities = 11/23 (47%), Positives = 13/23 (55%)
Frame = -3
Query: 54 ITFFLFFILLCFIFVFFFTFFIF 76
+ FF F F+F FFF FF F
Sbjct: 323 LCFFYFXXFFFFVFFFFFFFFFF 255
Score = 27.7 bits (60), Expect = 9.6
Identities = 12/25 (48%), Positives = 12/25 (48%)
Frame = -2
Query: 52 VVITFFLFFILLCFIFVFFFTFFIF 76
V F FF F F FFF FF F
Sbjct: 321 VFFLFXXFFFFCFFFFFFFFFFFFF 247
>BG363379 similar to GP|2224419|dbj|B ORF67 {Chlorella vulgaris}, partial
(26%)
Length = 360
Score = 30.8 bits (68), Expect = 1.1
Identities = 12/25 (48%), Positives = 13/25 (52%)
Frame = -2
Query: 52 VVITFFLFFILLCFIFVFFFTFFIF 76
+ FF FF F F FFF FF F
Sbjct: 203 IXXXFFFFFFFFFFFFFFFFFFFFF 129
Score = 28.9 bits (63), Expect = 4.3
Identities = 11/20 (55%), Positives = 12/20 (60%)
Frame = -2
Query: 56 FFLFFILLCFIFVFFFTFFI 75
FF FF F F FFF FF+
Sbjct: 173 FFFFFFFFFFFFFFFFFFFL 114
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.337 0.146 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,326,759
Number of Sequences: 63676
Number of extensions: 250242
Number of successful extensions: 6061
Number of sequences better than 10.0: 194
Number of HSP's better than 10.0 without gapping: 5241
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5849
length of query: 400
length of database: 12,639,632
effective HSP length: 99
effective length of query: 301
effective length of database: 6,335,708
effective search space: 1907048108
effective search space used: 1907048108
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146791.3


