

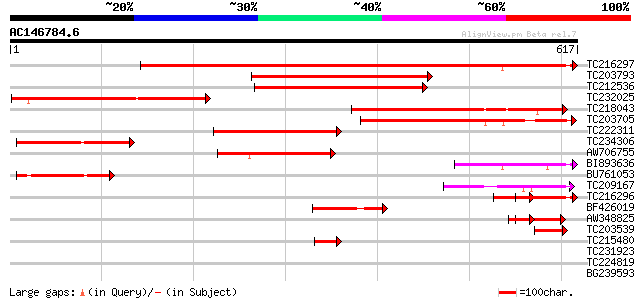
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146784.6 - phase: 0
(617 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216297 similar to UP|Q93X09 (Q93X09) Vacuolar sorting receptor... 763 0.0
TC203793 similar to PIR|T02602|T02602 vacuolar sorting receptor ... 330 1e-90
TC212536 homologue to UP|P93484 (P93484) BP-80 vacuolar sorting ... 310 9e-85
TC232025 similar to UP|P93484 (P93484) BP-80 vacuolar sorting re... 280 1e-75
TC218043 similar to UP|O49438 (O49438) Vacuolar sorting receptor... 254 8e-68
TC203705 similar to UP|P93484 (P93484) BP-80 vacuolar sorting re... 243 2e-64
TC222311 similar to UP|Q9FYH7 (Q9FYH7) F17F8.23, partial (21%) 221 7e-58
TC234306 homologue to UP|Q93X09 (Q93X09) Vacuolar sorting recept... 220 2e-57
AW706755 similar to GP|15487292|dbj vacuolar sorting receptor {V... 147 1e-35
BI893636 similar to GP|15487292|db vacuolar sorting receptor {Vi... 114 1e-25
BU761053 similar to GP|22530930|gb| vacuolar sorting receptor-li... 100 3e-21
TC209167 similar to UP|P93484 (P93484) BP-80 vacuolar sorting re... 92 5e-19
TC216296 similar to UP|Q93X09 (Q93X09) Vacuolar sorting receptor... 80 3e-15
BF426019 68 1e-11
AW348825 similar to GP|15487292|dbj vacuolar sorting receptor {V... 59 5e-09
TC203539 similar to UP|Q9SDR8 (Q9SDR8) Vacuolar sorting receptor... 55 7e-08
TC215480 similar to GB|BAA96951.1|8777361|AB019233 protein carbo... 43 4e-04
TC231923 weakly similar to UP|Q6QLL5 (Q6QLL5) WAK-like kinase, p... 33 0.49
TC224819 similar to GB|AAO39889.1|28372814|BT003661 At5g09570 {A... 29 5.4
BG239593 29 5.4
>TC216297 similar to UP|Q93X09 (Q93X09) Vacuolar sorting receptor, partial
(78%)
Length = 1641
Score = 763 bits (1970), Expect = 0.0
Identities = 359/483 (74%), Positives = 406/483 (83%), Gaps = 8/483 (1%)
Frame = +1
Query: 143 DYIEKINIPSALISKSLGDRIKKALSDGEMVHINLDWREALPHPDDRVEYELWTNSNDEC 202
DY++KI+IPSALISKSLGD IK+ALSDGEMV+INLDWRE+LPHPDDRVEYELWTNSNDEC
Sbjct: 1 DYVDKISIPSALISKSLGDSIKQALSDGEMVNINLDWRESLPHPDDRVEYELWTNSNDEC 180
Query: 203 GPKCDNQINFVKSFKGAAQLLEKKGFTQFTPHYITWYCPKEFLLSRRCKSQCINHGRYCA 262
GPKCD+ INF+K FKG AQ LEKKGFTQFTP YITW+CP+ FLLS++CKSQCIN+GRYCA
Sbjct: 181 GPKCDSLINFLKDFKGVAQQLEKKGFTQFTPRYITWFCPEAFLLSKQCKSQCINNGRYCA 360
Query: 263 PDPEQDFNKGYDGKDVVVQNLRQACFFKVANESGRPWQWWDYVTDFSIRCPMKEKKYTEE 322
PDPEQDF++GYDGKDVVVQNLRQACF+KVANESG+PWQWWDYVTDF+IRCPMKE KY+EE
Sbjct: 361 PDPEQDFSRGYDGKDVVVQNLRQACFYKVANESGKPWQWWDYVTDFAIRCPMKENKYSEE 540
Query: 323 CSDEVIKSLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKESRGDVTILPTLVINNRQ 382
CSD+VIKSLG DLKKIKDCVGDP ADVENPVLKAEQ+AQIG+ SRGDVTILPTLVINNRQ
Sbjct: 541 CSDQVIKSLGADLKKIKDCVGDPHADVENPVLKAEQDAQIGQGSRGDVTILPTLVINNRQ 720
Query: 383 YRGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGCWKEKSSNITACRDTFR 442
YRGKLSRP+VLKA+C+G+ ETTEPSICLT D+ETNECLENNGGCW++KSSNITACRDTFR
Sbjct: 721 YRGKLSRPSVLKAICSGYLETTEPSICLTSDLETNECLENNGGCWQDKSSNITACRDTFR 900
Query: 443 GRVCVCPVVNNIKFVGDGYTHCEASGTLSCEFNNGGCWKASHGGRLYSACHDDYRKGCEC 502
GRVC CP+V N+KF GDGYTHCEASG+LSCEFNNGGCWK + GGR YSAC DDYRKGC C
Sbjct: 901 GRVCECPIVQNVKFFGDGYTHCEASGSLSCEFNNGGCWKGAQGGRAYSACLDDYRKGCTC 1080
Query: 503 PSGFRGDGVRSCEDCFNIFRILTMLQILMNAK--------KSRLASVHNANAKIPLGVMS 554
P GFRGDGV+SCED + K K + ++ G S
Sbjct: 1081PPGFRGDGVQSCEDIDECNEKTSCQCPGCKCKNTWGSYECKCKSGLFYSRENDTCFGEYS 1260
Query: 555 ASAIAVCSTHEKMTRVLSGGYAFYKYRIQRYMDTEIRAIMAQYMPLDNQPLIIPNPNQVH 614
AS + + + ++GGYAFYKYRIQRYMD+EIR IMAQYMPLD+QP + NQVH
Sbjct: 1261ASVLNIWVIILVLVVAVAGGYAFYKYRIQRYMDSEIRTIMAQYMPLDSQPDV---SNQVH 1431
Query: 615 HDI 617
H+I
Sbjct: 1432HNI 1440
>TC203793 similar to PIR|T02602|T02602 vacuolar sorting receptor protein
homolog At2g14740 - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (31%)
Length = 711
Score = 330 bits (846), Expect = 1e-90
Identities = 144/197 (73%), Positives = 177/197 (89%)
Frame = +2
Query: 264 DPEQDFNKGYDGKDVVVQNLRQACFFKVANESGRPWQWWDYVTDFSIRCPMKEKKYTEEC 323
DPEQDF+ GYDGKDVV++NLRQ C FKVANE+ +PW WWDYVTDF IRCPMKEKKY ++C
Sbjct: 11 DPEQDFSTGYDGKDVVIENLRQLCVFKVANETKKPWVWWDYVTDFQIRCPMKEKKYNKKC 190
Query: 324 SDEVIKSLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKESRGDVTILPTLVINNRQY 383
+D VI+SLG+D+KKI+ C+GDP AD ENPVLK EQ+AQ+GK SRGDVTILPTLV+NNRQY
Sbjct: 191 ADAVIESLGLDIKKIERCMGDPNADSENPVLKEEQDAQVGKGSRGDVTILPTLVVNNRQY 370
Query: 384 RGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGCWKEKSSNITACRDTFRG 443
RGKL + AV+KA+CAGF+ETTEP++CL+ D+ETNECLENNGGCW++K +NITAC+DTFRG
Sbjct: 371 RGKLEKGAVMKAICAGFEETTEPAVCLSSDVETNECLENNGGCWQDKVANITACKDTFRG 550
Query: 444 RVCVCPVVNNIKFVGDG 460
RVC CP+V+ ++F G+G
Sbjct: 551 RVCECPLVDGVQFKGEG 601
>TC212536 homologue to UP|P93484 (P93484) BP-80 vacuolar sorting receptor,
partial (30%)
Length = 567
Score = 310 bits (795), Expect = 9e-85
Identities = 136/188 (72%), Positives = 167/188 (88%)
Frame = +3
Query: 267 QDFNKGYDGKDVVVQNLRQACFFKVANESGRPWQWWDYVTDFSIRCPMKEKKYTEECSDE 326
QDF GYDGKDVV++NLRQ C FKVANE+ +PW WWDYVTDF IRCPMKEKKY +EC++
Sbjct: 3 QDFTTGYDGKDVVIENLRQLCVFKVANETEKPWVWWDYVTDFQIRCPMKEKKYNKECANA 182
Query: 327 VIKSLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKESRGDVTILPTLVINNRQYRGK 386
VIKSLG++ +KI+ C+GDP AD +NPVLK EQ+AQIGK SRGDVTILPTLV+NNRQYRGK
Sbjct: 183 VIKSLGLNTEKIEKCMGDPDADTDNPVLKEEQDAQIGKGSRGDVTILPTLVVNNRQYRGK 362
Query: 387 LSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGCWKEKSSNITACRDTFRGRVC 446
L + AVLKA+C+GF+ETTEP++CL+ D+ETNECL NNGGCW++K++NITAC+DTFRGRVC
Sbjct: 363 LEKGAVLKAICSGFEETTEPTVCLSSDVETNECLTNNGGCWQDKAANITACKDTFRGRVC 542
Query: 447 VCPVVNNI 454
CP+V+ +
Sbjct: 543 ECPLVDGV 566
>TC232025 similar to UP|P93484 (P93484) BP-80 vacuolar sorting receptor,
partial (31%)
Length = 832
Score = 280 bits (716), Expect = 1e-75
Identities = 133/222 (59%), Positives = 168/222 (74%), Gaps = 6/222 (2%)
Frame = +1
Query: 3 IKLNPLLLCVLVLFLEC------CFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYG 56
+KL LCV + FL +F+VEKNSL +TSP ++KG+++ AIGNFG+PQYG
Sbjct: 169 MKLRRSSLCVFLGFLALFLSPPPSMAKFVVEKNSLTVTSPDNIKGTHDSAIGNFGIPQYG 348
Query: 57 GTLVGSVVYPNVNQKGCKNFTDFSASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAA 116
G++ G+V+YP N+KGCK F ++ SF S PG PT VL+DRG+C+F LK WNAQ GA+
Sbjct: 349 GSMAGNVLYPKDNKKGCKEFDEYGISFKSKPGALPTIVLLDRGNCFFALKVWNAQKAGAS 528
Query: 117 AILVADDREETLITMDTPEEGNVVNDDYIEKINIPSALISKSLGDRIKKALSDGEMVHIN 176
A+LV+DD EE LITMDTPEE + YIE I IPSALI KS G+++K A+S+G+MV++N
Sbjct: 529 AVLVSDDIEEKLITMDTPEEDG-SSAKYIENITIPSALIEKSFGEKLKVAISNGDMVNVN 705
Query: 177 LDWREALPHPDDRVEYELWTNSNDECGPKCDNQINFVKSFKG 218
LDWREA+PHPDDRVEYELWTNSNDECG KCD + FVK FKG
Sbjct: 706 LDWREAVPHPDDRVEYELWTNSNDECGVKCDMLMEFVKDFKG 831
>TC218043 similar to UP|O49438 (O49438) Vacuolar sorting receptor-like
protein, partial (36%)
Length = 1124
Score = 254 bits (649), Expect = 8e-68
Identities = 127/247 (51%), Positives = 160/247 (64%), Gaps = 12/247 (4%)
Frame = +2
Query: 373 LPTLVINNRQYRGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGCWKEKSS 432
LPTLVINN QYRGKL R AVLKA+C+GF+ETTEPS+CL+ D+ETNECLE NGGCW++K +
Sbjct: 2 LPTLVINNVQYRGKLERTAVLKAVCSGFKETTEPSVCLSGDVETNECLERNGGCWQDKHA 181
Query: 433 NITACRDTFRGRVCVCPVVNNIKFVGDGYTHCEASGTLSCEFNNGGCWKASHGGRLYSAC 492
NITAC+DTFRGRVC CPVVN +++ GDGYT CEA G C NNGGCW + G +SAC
Sbjct: 182 NITACKDTFRGRVCECPVVNGVQYKGDGYTTCEAFGPARCSINNGGCWSETKKGLTFSAC 361
Query: 493 HDDYRKGCECPSGFRGDGVRSCEDCFNIFRILTMLQILMNAKKSRLASVHNANAKIPLGV 552
D GC+CP GFRGDG CED + + + Q + K+ S ++ K L
Sbjct: 362 SDSKVNGCQCPVGFRGDGTNKCED-VDECKERSACQCDGCSCKNTWGS-YDCKCKGNLLY 535
Query: 553 MSASAIAVCSTHEKMTRVLS------------GGYAFYKYRIQRYMDTEIRAIMAQYMPL 600
+ + + K R L+ GY FYKYR++ YMD+EI AIM+QYMPL
Sbjct: 536 IKEQDACIERSESKFGRFLAFVVIAVVVGAGLAGYVFYKYRLRSYMDSEIMAIMSQYMPL 715
Query: 601 DNQPLII 607
D Q ++
Sbjct: 716 DQQNNVV 736
>TC203705 similar to UP|P93484 (P93484) BP-80 vacuolar sorting receptor,
partial (38%)
Length = 1045
Score = 243 bits (619), Expect = 2e-64
Identities = 124/258 (48%), Positives = 161/258 (62%), Gaps = 23/258 (8%)
Frame = +1
Query: 382 QYRGKLSRPAVLKAMCAGFQETTEPSICLTPDMETNECLENNGGCWKEKSSNITACRDTF 441
QYRGKL + AV+KA+CAGF+ETTEP++CL+ D+ETNECLENNGGCW++K +NITAC+DTF
Sbjct: 1 QYRGKLEKGAVMKAICAGFEETTEPAVCLSSDVETNECLENNGGCWQDKVANITACKDTF 180
Query: 442 RGRVCVCPVVNNIKFVGDGYTHCEASGTLSCEFNNGGCWKASHGGRLYSACHDDYRKGCE 501
RGRVC CP+V+ ++F GDGYT C ASG C+ NNGGCW + G YSAC DD C+
Sbjct: 181 RGRVCECPLVDGVQFKGDGYTICAASGPGRCKINNGGCWHEARNGHAYSACSDDGGVKCQ 360
Query: 502 CPSGFRGDGVRSCED----------------CFNIFRI--LTMLQILMNAKK-----SRL 538
CP+GF+GDGV++CED C N + T L+ + S+
Sbjct: 361 CPAGFKGDGVKNCEDIDECKEKKACQCPECSCKNTWGSYDCTCSGDLLYIRDHDTCISKT 540
Query: 539 ASVHNANAKIPLGVMSASAIAVCSTHEKMTRVLSGGYAFYKYRIQRYMDTEIRAIMAQYM 598
AS +A V+ + +G Y YKYRI+ YMD+EIRAIMAQYM
Sbjct: 541 ASQEGRSAWAAFWVILVGLVVAA----------AGAYLVYKYRIRSYMDSEIRAIMAQYM 690
Query: 599 PLDNQPLIIPNPNQVHHD 616
PLD+Q ++ N VH +
Sbjct: 691 PLDSQGEVV---NHVHEE 735
>TC222311 similar to UP|Q9FYH7 (Q9FYH7) F17F8.23, partial (21%)
Length = 421
Score = 221 bits (563), Expect = 7e-58
Identities = 94/140 (67%), Positives = 117/140 (83%)
Frame = +1
Query: 222 LLEKKGFTQFTPHYITWYCPKEFLLSRRCKSQCINHGRYCAPDPEQDFNKGYDGKDVVVQ 281
+LE+ G+T FTPHYITW+CP F+LS +CKSQCINHGRYCAPDPE+DF +GY+GKDVV +
Sbjct: 1 ILERGGYTLFTPHYITWFCPPPFILSSQCKSQCINHGRYCAPDPEKDFGEGYEGKDVVYE 180
Query: 282 NLRQACFFKVANESGRPWQWWDYVTDFSIRCPMKEKKYTEECSDEVIKSLGVDLKKIKDC 341
NLRQ C +VANES R W WWDYVTDF +RC MKEK+Y+++C++EV+KSL + + KIK C
Sbjct: 181 NLRQLCVHRVANESNRSWVWWDYVTDFHVRCSMKEKRYSKDCAEEVMKSLDLPVDKIKKC 360
Query: 342 VGDPLADVENPVLKAEQEAQ 361
+GDP ADVEN VLK EQ+ Q
Sbjct: 361 MGDPEADVENEVLKNEQQVQ 420
>TC234306 homologue to UP|Q93X09 (Q93X09) Vacuolar sorting receptor, partial
(21%)
Length = 639
Score = 220 bits (560), Expect = 2e-57
Identities = 105/129 (81%), Positives = 115/129 (88%)
Frame = +1
Query: 8 LLLCVLVLFLECCFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYGGTLVGSVVYPN 67
LLLCV VL L CC GRF+VEKNSL++TSPKSLKG+YECAIGNFGVP+YGGTLVGSV+YP
Sbjct: 259 LLLCVGVLLLRCCVGRFVVEKNSLKVTSPKSLKGTYECAIGNFGVPKYGGTLVGSVLYPK 438
Query: 68 VNQKGCKNFTDFSASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGGAAAILVADDREET 127
VNQKGC NF+D +F S PG PTF+LVDRGDCYFTLKAWNAQNGGAAAILVADD+ ET
Sbjct: 439 VNQKGCTNFSD--VNFQSKPGGSPTFLLVDRGDCYFTLKAWNAQNGGAAAILVADDKAET 612
Query: 128 LITMDTPEE 136
LITMDTPEE
Sbjct: 613 LITMDTPEE 639
>AW706755 similar to GP|15487292|dbj vacuolar sorting receptor {Vigna mungo},
partial (12%)
Length = 421
Score = 147 bits (371), Expect = 1e-35
Identities = 71/132 (53%), Positives = 89/132 (66%), Gaps = 4/132 (3%)
Frame = +2
Query: 227 GFTQFTPHYITWYCPKEFLLSRRCKSQCINHGR----YCAPDPEQDFNKGYDGKDVVVQN 282
GFT FT ++T + P+ F L+++ + + +P K + V N
Sbjct: 2 GFTNFTLGFLTGFGPEPFFLTKQANFNALTMEKGLCVLISPTKYSQIEGSISEKILFVLN 181
Query: 283 LRQACFFKVANESGRPWQWWDYVTDFSIRCPMKEKKYTEECSDEVIKSLGVDLKKIKDCV 342
LR ACF+KVANESG+PWQWWDYVTDFSIRCPMKE K++EEC D+VIKSLGVDL+K KDCV
Sbjct: 182 LRPACFYKVANESGKPWQWWDYVTDFSIRCPMKENKFSEECVDQVIKSLGVDLEKNKDCV 361
Query: 343 GDPLADVENPVL 354
GDP AD+ N VL
Sbjct: 362 GDPHADI*NAVL 397
>BI893636 similar to GP|15487292|db vacuolar sorting receptor {Vigna mungo},
partial (22%)
Length = 437
Score = 114 bits (286), Expect = 1e-25
Identities = 68/146 (46%), Positives = 82/146 (55%), Gaps = 13/146 (8%)
Frame = +2
Query: 485 GGRLYSACHDDYRKGCECPSGFRGDGVRSCEDCFNIFRILTMLQILMNAK--------KS 536
GGR YSAC DDYRKGC CP GFRGDGV+SCED + K K
Sbjct: 2 GGRAYSACLDDYRKGCTCPPGFRGDGVQSCEDIDECNEKTSCQCPGCKCKNTWGSYECKC 181
Query: 537 RLASVHNANAKIPLGVMSASAIAVCSTHEKMTRVLSGGYAFYKYRIQ-----RYMDTEIR 591
+ ++ G SAS + + + ++GGYAFYKYRIQ RYMD+EIR
Sbjct: 182 KSGLFYSRENDTCFGEYSASVLNIWVIILVLVVAVAGGYAFYKYRIQLIYVQRYMDSEIR 361
Query: 592 AIMAQYMPLDNQPLIIPNPNQVHHDI 617
IMAQYMPLD+QP + NQVHH+I
Sbjct: 362 TIMAQYMPLDSQPDV---SNQVHHNI 430
>BU761053 similar to GP|22530930|gb| vacuolar sorting receptor-like protein
{Arabidopsis thaliana}, partial (12%)
Length = 446
Score = 99.8 bits (247), Expect = 3e-21
Identities = 52/107 (48%), Positives = 68/107 (62%)
Frame = +1
Query: 8 LLLCVLVLFLECCFGRFLVEKNSLRITSPKSLKGSYECAIGNFGVPQYGGTLVGSVVYPN 67
LLL V+V+ ++ RF+VEK+S+ + SP LK + AIGNFG+P YGG +VGSVVYP
Sbjct: 139 LLLLVVVVVVDA---RFVVEKSSITVLSPHKLKAKRDGAIGNFGLPDYGGFIVGSVVYPA 309
Query: 68 VNQKGCKNFTDFSASFHSMPGNFPTFVLVDRGDCYFTLKAWNAQNGG 114
GC+NF + PT VL+DRG+CYF LK W+AQ G
Sbjct: 310 KGSHGCENFEGDKP--FKIQSYRPTIVLLDRGECYFALKVWHAQLAG 444
>TC209167 similar to UP|P93484 (P93484) BP-80 vacuolar sorting receptor,
partial (25%)
Length = 783
Score = 92.4 bits (228), Expect = 5e-19
Identities = 54/167 (32%), Positives = 79/167 (46%), Gaps = 25/167 (14%)
Frame = +1
Query: 473 EFNNGGCWKASHGGRLYSACHDDYRKGCECPSGFRGDGVRSCEDCFNIFRILTMLQILMN 532
+ NNGGCW + G +SAC D+ C+CP+GFRGDGV++C+D +
Sbjct: 10 KINNGGCWHDAQNGHAFSACLDNGGVKCQCPTGFRGDGVKNCQD-------------IDE 150
Query: 533 AKKSRLASVHNANAKIPLGVMSASA---IAVCSTHEK----------------------M 567
K+ + + + K G S + H+ +
Sbjct: 151 CKEKKACQCPDCSCKNTWGSYDCSCSGDLLYMRDHDTCISKTGSQGRSTWAAFWLILLGV 330
Query: 568 TRVLSGGYAFYKYRIQRYMDTEIRAIMAQYMPLDNQPLIIPNPNQVH 614
+ G + YKYRI++YMD+EIRAIMAQYMPLD+Q + PN V+
Sbjct: 331 VMIAGGAFLVYKYRIRQYMDSEIRAIMAQYMPLDSQAEV---PNHVN 462
>TC216296 similar to UP|Q93X09 (Q93X09) Vacuolar sorting receptor, partial
(18%)
Length = 648
Score = 80.1 bits (196), Expect = 3e-15
Identities = 41/67 (61%), Positives = 49/67 (72%)
Frame = +3
Query: 551 GVMSASAIAVCSTHEKMTRVLSGGYAFYKYRIQRYMDTEIRAIMAQYMPLDNQPLIIPNP 610
G SAS + + + ++GGYAFYKYRIQRYMD+EIRAIMAQYMPLDNQP +
Sbjct: 141 GEYSASVLNIWVIILVLVVAVAGGYAFYKYRIQRYMDSEIRAIMAQYMPLDNQPEV---S 311
Query: 611 NQVHHDI 617
NQVHH+I
Sbjct: 312 NQVHHNI 332
Score = 47.0 bits (110), Expect = 3e-05
Identities = 25/45 (55%), Positives = 32/45 (70%)
Frame = +1
Query: 527 LQILMNAKKSRLASVHNANAKIPLGVMSASAIAVCSTHEKMTRVL 571
++ILM+AK+ + + ANAK P V SA+A VCST EKMTRVL
Sbjct: 7 VKILMSAKRKQPVNAQVANAKTPGEVTSANAKVVCSTREKMTRVL 141
>BF426019
Length = 408
Score = 68.2 bits (165), Expect = 1e-11
Identities = 36/82 (43%), Positives = 57/82 (68%)
Frame = +3
Query: 330 SLGVDLKKIKDCVGDPLADVENPVLKAEQEAQIGKESRGDVTILPTLVINNRQYRGKLSR 389
S G++ +KI+ C+GDP AD +NPVLK EQ+AQ+ + G + ++VI YR L++
Sbjct: 69 SAGLNTEKIEKCMGDPDADTDNPVLKEEQDAQVLSFNNGYLVHCISVVI----YR-CLTK 233
Query: 390 PAVLKAMCAGFQETTEPSICLT 411
+ A+C+GF+ETTEP++CL+
Sbjct: 234 LVFIIAICSGFEETTEPAVCLS 299
>AW348825 similar to GP|15487292|dbj vacuolar sorting receptor {Vigna mungo},
partial (15%)
Length = 475
Score = 59.3 bits (142), Expect = 5e-09
Identities = 30/54 (55%), Positives = 36/54 (66%)
Frame = -2
Query: 551 GVMSASAIAVCSTHEKMTRVLSGGYAFYKYRIQRYMDTEIRAIMAQYMPLDNQP 604
G SAS + + + ++GGYAFYK RIQRYMD+EIR IMAQ MPLD P
Sbjct: 345 GEYSASVLNIWVIILVLVVAVAGGYAFYKXRIQRYMDSEIRTIMAQXMPLDXXP 184
Score = 44.3 bits (103), Expect = 2e-04
Identities = 22/28 (78%), Positives = 23/28 (81%)
Frame = -3
Query: 544 ANAKIPLGVMSASAIAVCSTHEKMTRVL 571
ANAK P GV SA+A VCSTHEKMTRVL
Sbjct: 428 ANAKTPGGVTSANAKVVCSTHEKMTRVL 345
>TC203539 similar to UP|Q9SDR8 (Q9SDR8) Vacuolar sorting receptor protein
(Fragment), partial (66%)
Length = 648
Score = 55.5 bits (132), Expect = 7e-08
Identities = 25/36 (69%), Positives = 30/36 (82%)
Frame = +1
Query: 572 SGGYAFYKYRIQRYMDTEIRAIMAQYMPLDNQPLII 607
+G Y YKYRI+ YMD+EIRAIMAQYMPLD+Q I+
Sbjct: 199 AGAYLVYKYRIRSYMDSEIRAIMAQYMPLDSQGEIV 306
>TC215480 similar to GB|BAA96951.1|8777361|AB019233 protein carboxyl
methylase-like {Arabidopsis thaliana;} , partial (21%)
Length = 955
Score = 43.1 bits (100), Expect = 4e-04
Identities = 18/30 (60%), Positives = 25/30 (83%)
Frame = +1
Query: 332 GVDLKKIKDCVGDPLADVENPVLKAEQEAQ 361
G++ +KI+ C+GDP AD +NPVLK EQ+AQ
Sbjct: 598 GLNTEKIEKCMGDPDADTDNPVLKEEQDAQ 687
>TC231923 weakly similar to UP|Q6QLL5 (Q6QLL5) WAK-like kinase, partial (24%)
Length = 600
Score = 32.7 bits (73), Expect = 0.49
Identities = 18/50 (36%), Positives = 23/50 (46%)
Frame = +3
Query: 468 GTLSCEFNNGGCWKASHGGRLYSACHDDYRKGCECPSGFRGDGVRSCEDC 517
G SC NN C + +HGG +R C C GF GDG + + C
Sbjct: 132 GNCSCS-NNASCTEVNHGGGKLG-----FR--CRCDEGFVGDGFKDGDGC 257
>TC224819 similar to GB|AAO39889.1|28372814|BT003661 At5g09570 {Arabidopsis
thaliana;} , partial (60%)
Length = 751
Score = 29.3 bits (64), Expect = 5.4
Identities = 17/44 (38%), Positives = 21/44 (47%)
Frame = +3
Query: 460 GYTHCEASGTLSCEFNNGGCWKASHGGRLYSACHDDYRKGCECP 503
G+ H + SGT C GC SHG +S + YR C CP
Sbjct: 255 GFWHRKCSGTQGC-----GC---SHGSSYHST*NSCYRGCCCCP 362
>BG239593
Length = 595
Score = 29.3 bits (64), Expect = 5.4
Identities = 14/40 (35%), Positives = 21/40 (52%)
Frame = +2
Query: 570 VLSGGYAFYKYRIQRYMDTEIRAIMAQYMPLDNQPLIIPN 609
+LS G+ F + IQR++D R +PL +IPN
Sbjct: 56 ILSSGHTFDRSSIQRWLDAGHRTCPITKLPLPAHSSLIPN 175
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.138 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,051,766
Number of Sequences: 63676
Number of extensions: 510657
Number of successful extensions: 2487
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 2447
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2475
length of query: 617
length of database: 12,639,632
effective HSP length: 103
effective length of query: 514
effective length of database: 6,081,004
effective search space: 3125636056
effective search space used: 3125636056
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146784.6


