

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146784.3 - phase: 0
(348 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
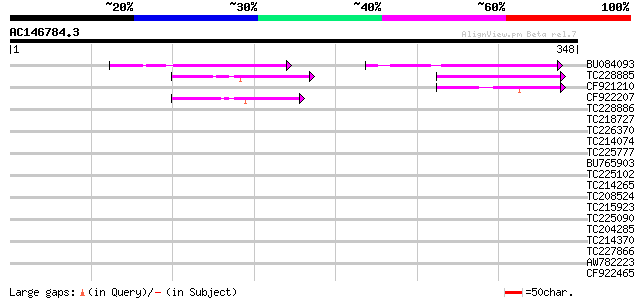
Score E
Sequences producing significant alignments: (bits) Value
BU084093 70 1e-12
TC228885 48 6e-06
CF921210 43 2e-04
CF922207 42 4e-04
TC228886 37 0.017
TC218727 similar to UP|Q8LSC5 (Q8LSC5) Geranylgeranyl pyrophosph... 35 0.066
TC226370 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pr... 34 0.086
TC214074 homologue to UP|Q6WV72 (Q6WV72) Histone H4, complete 34 0.11
TC225777 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 ... 33 0.19
BU765903 33 0.25
TC225102 homologue to UP|Q6WV72 (Q6WV72) Histone H4, complete 33 0.25
TC214265 UP|PRP3_SOYBN (P15642) Repetitive proline-rich cell wal... 33 0.25
TC208524 weakly similar to UP|Q6SPE9 (Q6SPE9) Atherin, partial (7%) 33 0.25
TC215923 similar to GB|AAS68109.1|45592910|BT011785 At2g38360 {A... 32 0.33
TC225090 homologue to UP|Q6WV72 (Q6WV72) Histone H4, complete 32 0.43
TC204285 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 32 0.43
TC214370 homologue to UP|Q6WV72 (Q6WV72) Histone H4, complete 32 0.43
TC227866 similar to UP|P2B1_DROME (P48456) Serine/threonine prot... 32 0.56
AW782223 32 0.56
CF922465 31 0.73
>BU084093
Length = 421
Score = 70.5 bits (171), Expect = 1e-12
Identities = 37/112 (33%), Positives = 55/112 (49%)
Frame = -1
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLK 121
+P++P Q PQ P R + + + F P+ + P+PM Y DLLP L+
Sbjct: 340 QPKAPAQREAPQVPTPNTTRP-VGNSNTTRNFPPR----PFQEFTPLPMTYEDLLPSLIA 176
Query: 122 KNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIE 173
+L P + P WY P+ C +H G PGH E+C LK +VQ L++
Sbjct: 175 NHLAVVTPGRVLQPPFPKWYDPNATCKYHGGVPGHSVEKCLALKYKVQHLMD 20
Score = 62.4 bits (150), Expect = 3e-10
Identities = 35/121 (28%), Positives = 57/121 (46%)
Frame = -1
Query: 219 PSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPIPMKYGELFPY 278
P+Q++ P +P N R G + + +P Q+ F P+PM Y +L P
Sbjct: 328 PAQREAPQ-------VPTPNTTRPVGNSNTTRNFPPRPFQE-----FTPLPMTYEDLLPS 185
Query: 279 LLERNLVQTRPPPPIPKKLPARWRPDLFCVFHQGAQGHDVERCFSLKIEVQKLIEDDLIP 338
L+ +L P + P + P+ C +H G GH VE+C +LK +VQ L++ +
Sbjct: 184 LIANHLAVVTPGRVLQPPFPKWYDPNATCKYHGGVPGHSVEKCLALKYKVQHLMDAGWLT 5
Query: 339 F 339
F
Sbjct: 4 F 2
>TC228885
Length = 901
Score = 48.1 bits (113), Expect = 6e-06
Identities = 32/91 (35%), Positives = 47/91 (51%), Gaps = 3/91 (3%)
Frame = -3
Query: 100 QKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPW---YRPDLNCVFHQGAPGH 156
+K + PIP+ YADLLP LL ++V + L +V PP+ Y + C H APG
Sbjct: 632 RKPVEFTPIPVSYADLLPYLLDNSMV-AITLAKVHQ--PPFLREYDSNAMCACHGEAPGR 462
Query: 157 DTEQCYPLKEEVQKLIENNVWSFDDQDIKVL 187
E LK +VQ LI+ F++ + +L
Sbjct: 461 SIEHYRALKRKVQGLIDAGWLKFEENRV*IL 369
Score = 47.4 bits (111), Expect = 1e-05
Identities = 25/79 (31%), Positives = 39/79 (48%)
Frame = -3
Query: 263 IKFDPIPMKYGELFPYLLERNLVQTRPPPPIPKKLPARWRPDLFCVFHQGAQGHDVERCF 322
++F PIP+ Y +L PYLL+ ++V + + C H A G +E
Sbjct: 623 VEFTPIPVSYADLLPYLLDNSMVAITLAKVHQPPFLREYDSNAMCACHGEAPGRSIEHYR 444
Query: 323 SLKIEVQKLIEDDLIPFEE 341
+LK +VQ LI+ + FEE
Sbjct: 443 ALKRKVQGLIDAGWLKFEE 387
>CF921210
Length = 790
Score = 43.1 bits (100), Expect = 2e-04
Identities = 28/84 (33%), Positives = 42/84 (49%), Gaps = 5/84 (5%)
Frame = +2
Query: 263 IKFDPIPMKYGELFPYLLERNLVQTRPPPPIPKKLPARWRPDLFCVFHQ-----GAQGHD 317
++F PIP+ Y +L YLL+ ++V L +P LF + GA H
Sbjct: 335 VEFTPIPVSYADLLSYLLDNSMVAIT--------LAKVHQPPLF*GYDSNATCGGALRHS 490
Query: 318 VERCFSLKIEVQKLIEDDLIPFEE 341
+E C +LK +VQ LI+ + FEE
Sbjct: 491 IEHCRALKRKVQGLIDAGWLKFEE 562
Score = 40.0 bits (92), Expect = 0.002
Identities = 31/91 (34%), Positives = 45/91 (49%), Gaps = 3/91 (3%)
Frame = +2
Query: 100 QKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPP---WYRPDLNCVFHQGAPGH 156
+K + PIP+ YADLL LL ++V + L +V PP Y + C GA H
Sbjct: 326 RKPVEFTPIPVSYADLLSYLLDNSMV-AITLAKVHQ--PPLF*GYDSNATC---GGALRH 487
Query: 157 DTEQCYPLKEEVQKLIENNVWSFDDQDIKVL 187
E C LK +VQ LI+ F++ + +L
Sbjct: 488 SIEHCRALKRKVQGLIDAGWLKFEENCV*IL 580
>CF922207
Length = 616
Score = 42.0 bits (97), Expect = 4e-04
Identities = 28/86 (32%), Positives = 42/86 (48%), Gaps = 4/86 (4%)
Frame = -1
Query: 100 QKASQCDP-IPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRP---DLNCVFHQGAPG 155
QKA + P +L+P L ++V P +VP PP++R + C +H GAPG
Sbjct: 580 QKACRVHPKFRCHMLNLIPYPLDNSMVAITPT-KVPQ--PPFFREYDSNATCAYHGGAPG 410
Query: 156 HDTEQCYPLKEEVQKLIENNVWSFDD 181
H E C K +V LI+ F++
Sbjct: 409 HSIEHCMTPKHKV*SLIDTG*LKFEE 332
Score = 40.8 bits (94), Expect = 0.001
Identities = 21/67 (31%), Positives = 32/67 (47%)
Frame = -1
Query: 275 LFPYLLERNLVQTRPPPPIPKKLPARWRPDLFCVFHQGAQGHDVERCFSLKIEVQKLIED 334
L PY L+ ++V P + + C +H GA GH +E C + K +V LI+
Sbjct: 532 LIPYPLDNSMVAITPTKVPQPPFFREYDSNATCAYHGGAPGHSIEHCMTPKHKV*SLIDT 353
Query: 335 DLIPFEE 341
+ FEE
Sbjct: 352 G*LKFEE 332
>TC228886
Length = 748
Score = 36.6 bits (83), Expect = 0.017
Identities = 15/35 (42%), Positives = 21/35 (59%)
Frame = +3
Query: 307 CVFHQGAQGHDVERCFSLKIEVQKLIEDDLIPFEE 341
C +H GA GH +E C + K +V LI+ + FEE
Sbjct: 171 CAYHGGASGHSIEHCMTPKHKV*SLIDTGWLKFEE 275
Score = 35.4 bits (80), Expect = 0.039
Identities = 19/61 (31%), Positives = 29/61 (47%), Gaps = 3/61 (4%)
Frame = +3
Query: 124 LVQTLPLPRVPNSLPPW---YRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFD 180
+ Q P P + + PP+ Y + C +H GA GH E C K +V LI+ F+
Sbjct: 93 ITQW*P*PLLGSLKPPFF*GYDSNATCAYHGGASGHSIEHCMTPKHKV*SLIDTGWLKFE 272
Query: 181 D 181
+
Sbjct: 273 E 275
>TC218727 similar to UP|Q8LSC5 (Q8LSC5) Geranylgeranyl pyrophosphate
synthase, partial (55%)
Length = 892
Score = 34.7 bits (78), Expect = 0.066
Identities = 25/78 (32%), Positives = 32/78 (40%)
Frame = -1
Query: 66 PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLV 125
P QP P P A R+ IP +Q P P N +P + L+P L+
Sbjct: 292 PTQPQLPATPPFGA-RE*IPASQAPHSPPPPNLYPPLVPPQALPYRSTSLIPQSLE---- 128
Query: 126 QTLPLPRVPNSLPPWYRP 143
PRV +LPPW P
Sbjct: 127 -----PRVTATLPPWTLP 89
>TC226370 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (43%)
Length = 1810
Score = 34.3 bits (77), Expect = 0.086
Identities = 22/61 (36%), Positives = 28/61 (45%), Gaps = 1/61 (1%)
Frame = +2
Query: 203 PITNVVQDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQ-PQFQQYQQQPRQQAP 261
P+ N Q Q+ PS QQY V V +P Q +QQ QQQP+QQ P
Sbjct: 731 PLPNPAVPQHPQNQYLPSDQQYRTPQHVAPQPTPSQVTPSPVQQFSHYQQQQQQPQQQQP 910
Query: 262 R 262
+
Sbjct: 911 Q 913
Score = 33.1 bits (74), Expect = 0.19
Identities = 78/325 (24%), Positives = 107/325 (32%), Gaps = 40/325 (12%)
Frame = +2
Query: 15 QTGNMDQLEQEVQELRGEVTTLR-----AEVEK-LASLVSSLMATNDPPLVQQRPQSPYQ 68
Q + LE+ VQE+ V LR AE +K LA L + ++ Q +
Sbjct: 365 QESRLKSLEKHVQEVHRSVQILRDKQELAETQKELAKLQLAQKESSSSSHSQSNEERSSP 544
Query: 69 PMCPQKP------RQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKK 122
P+K Q ++P PQ P QVQ +Q P P
Sbjct: 545 TTDPKKTDNASDANNQQLALALPHQIAPQPQPPAPQVQAQAQA-PAP------------- 682
Query: 123 NLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQ 182
N+ Q VP PP+Y P + P H Q P DQ
Sbjct: 683 NVTQ------VPQQ-PPYYMPPTPLP-NPAVPQHPQNQYLP----------------SDQ 790
Query: 183 DIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYP--DSHFVTTIL-----P 235
+ QH+AP + + V Q YQ Q + QQQ P + +L P
Sbjct: 791 QYRT---PQHVAPQPTPSQVTPSPVQQFSHYQQQQQQPQQQQPQQQQQWSQQVLPSQPPP 961
Query: 236 IMNVVR--NPGYQPQFQQYQ----------------QQPRQQAP---RIKFDPIPMKYGE 274
+ + VR +P P +Q Q Q P P + D IP YG
Sbjct: 962 MQSQVRPPSPNVYPPYQPNQATNPSPAETLPNSMAMQMPYSGVPPPGSSRTDAIPYGYGG 1141
Query: 275 LFPYLLERNLVQTRPPPPIPKKLPA 299
R + Q PP + PA
Sbjct: 1142-----AGRTVPQQPPPQQMKSSFPA 1201
>TC214074 homologue to UP|Q6WV72 (Q6WV72) Histone H4, complete
Length = 662
Score = 33.9 bits (76), Expect = 0.11
Identities = 20/50 (40%), Positives = 28/50 (56%), Gaps = 5/50 (10%)
Frame = -3
Query: 261 PRIKFDPIPMKY-----GELFPYLLERNLVQTRPPPPIPKKLPARWRPDL 305
P I+F P P++ G + P +L RN ++ R PP PK LP RPD+
Sbjct: 246 PLIRFTP-PLRAKRRIAGLVIP*MLSRNTLRCRFAPPFPKPLPPFPRPDI 100
>TC225777 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 protein
(Corresponding sequence ZK643.8), partial (7%)
Length = 762
Score = 33.1 bits (74), Expect = 0.19
Identities = 23/74 (31%), Positives = 27/74 (36%)
Frame = +3
Query: 190 QQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQF 249
QQHLAP + PI P + PQF Q QF
Sbjct: 225 QQHLAPQQLPPQPPIFQQTYPPFHLPQFHHHQPSL-----------------------QF 335
Query: 250 QQYQQQPRQQAPRI 263
QQ QQQP Q P++
Sbjct: 336 QQQQQQPPQPPPQL 377
Score = 27.7 bits (60), Expect = 8.0
Identities = 16/40 (40%), Positives = 21/40 (52%)
Frame = +3
Query: 57 PLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQ 96
P VQQ+ Q P Q + QQ P+Q Q+ PQ+ PQ
Sbjct: 159 PPVQQQQQPPQQQL------QQPPQQPPQQHLAPQQLPPQ 260
>BU765903
Length = 387
Score = 32.7 bits (73), Expect = 0.25
Identities = 24/87 (27%), Positives = 38/87 (43%), Gaps = 3/87 (3%)
Frame = +2
Query: 219 PSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPIP---MKYGEL 275
P Q P+ H T + R P + Q Q + R++AP + +P + L
Sbjct: 128 PPQTPNPNPHHTTLGWTHFSPFRTPARR-QIHQARLHRRRRAPLLLLPDLPHARLVQASL 304
Query: 276 FPYLLERNLVQTRPPPPIPKKLPARWR 302
P + R L + R P P+P++ RWR
Sbjct: 305 LP--IRRRLGRRRLPLPLPQRARPRWR 379
>TC225102 homologue to UP|Q6WV72 (Q6WV72) Histone H4, complete
Length = 1012
Score = 32.7 bits (73), Expect = 0.25
Identities = 15/37 (40%), Positives = 21/37 (56%)
Frame = -2
Query: 273 GELFPYLLERNLVQTRPPPPIPKKLPARWRPDLFCVF 309
G + P++L R+ PP+PK LP RPD+F F
Sbjct: 327 GFVIPWMLSRSTFLCLLAPPLPKPLPPFPRPDIFARF 217
>TC214265 UP|PRP3_SOYBN (P15642) Repetitive proline-rich cell wall protein 3
precursor, complete
Length = 849
Score = 32.7 bits (73), Expect = 0.25
Identities = 25/91 (27%), Positives = 44/91 (47%), Gaps = 7/91 (7%)
Frame = +3
Query: 60 QQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDP-------IPMKY 112
+++P++ YQP+ + +Q R + +N +F+PQNQ K + P P K
Sbjct: 141 RKQPRNQYQPLRLLQQNKQQLRFLLKKNNKQLRFLPQNQPPKHQRKKPPKHQHQNKPTKM 320
Query: 113 ADLLPILLKKNLVQTLPLPRVPNSLPPWYRP 143
A + L+ L+ L L +P L +Y+P
Sbjct: 321 ASFVSFLVL--LLAALIL--MPQGLATYYKP 401
>TC208524 weakly similar to UP|Q6SPE9 (Q6SPE9) Atherin, partial (7%)
Length = 544
Score = 32.7 bits (73), Expect = 0.25
Identities = 19/57 (33%), Positives = 26/57 (45%)
Frame = +2
Query: 36 LRAEVEKLASLVSSLMATNDPPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQK 92
L+ V+ A S PPLVQ +PQ QP +P+Q+ Q Q VP +
Sbjct: 329 LQTLVDANAKHFSGFSPLPPPPLVQPQPQPQPQPQPQPQPQQEQQPQQQQQGMVPSQ 499
>TC215923 similar to GB|AAS68109.1|45592910|BT011785 At2g38360 {Arabidopsis
thaliana;} , partial (76%)
Length = 1055
Score = 32.3 bits (72), Expect = 0.33
Identities = 24/69 (34%), Positives = 29/69 (41%)
Frame = +3
Query: 241 RNPGYQPQFQQYQQQPRQQAPRIKFDPIPMKYGELFPYLLERNLVQTRPPPPIPKKLPAR 300
R G QP ++ QQP Q P + P P G P + V R PP P+KL
Sbjct: 219 RGSGQQPGLPRFHQQPLQLPPP-RPRPAPPLVGARGPLSVLETRVLLRSHPPCPQKLLLL 395
Query: 301 WRPDLFCVF 309
R L C F
Sbjct: 396 PRQLLRCRF 422
>TC225090 homologue to UP|Q6WV72 (Q6WV72) Histone H4, complete
Length = 681
Score = 32.0 bits (71), Expect = 0.43
Identities = 15/37 (40%), Positives = 20/37 (53%)
Frame = -3
Query: 273 GELFPYLLERNLVQTRPPPPIPKKLPARWRPDLFCVF 309
G + P++L R+ PP PK LP RPD+F F
Sbjct: 256 GLVIPWMLSRSTFLCLLAPPFPKPLPPFPRPDIFARF 146
>TC204285 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (32%)
Length = 1539
Score = 32.0 bits (71), Expect = 0.43
Identities = 24/86 (27%), Positives = 38/86 (43%)
Frame = +1
Query: 54 NDPPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYA 113
N PPL + P+ P P P++P P++ P + +K +P + K P+P+
Sbjct: 616 NPPPLPPKPPKEPPTP--PKEPPPTPPKKPCPP--IFKKPLPP--IPKLPPLPPVPIYKK 777
Query: 114 DLLPILLKKNLVQTLPLPRVPNSLPP 139
L P+ L P+P LPP
Sbjct: 778 PLPPLPPLPKLPPLHPVPIYKKPLPP 855
>TC214370 homologue to UP|Q6WV72 (Q6WV72) Histone H4, complete
Length = 916
Score = 32.0 bits (71), Expect = 0.43
Identities = 19/50 (38%), Positives = 28/50 (56%), Gaps = 5/50 (10%)
Frame = -2
Query: 261 PRIKFDPIPMKY-----GELFPYLLERNLVQTRPPPPIPKKLPARWRPDL 305
P I+F P P++ G + P +L R+ ++ R PP PK LP RPD+
Sbjct: 330 PLIRFTP-PLRAKRRIAGLVIP*MLSRSTLRCRFAPPFPKPLPPFPRPDI 184
>TC227866 similar to UP|P2B1_DROME (P48456) Serine/threonine protein
phosphatase 2B catalytic subunit 1
(Calmodulin-dependent calcineurin A1 subunit) , partial
(3%)
Length = 1013
Score = 31.6 bits (70), Expect = 0.56
Identities = 15/42 (35%), Positives = 21/42 (49%)
Frame = +3
Query: 253 QQQPRQQAPRIKFDPIPMKYGELFPYLLERNLVQTRPPPPIP 294
QQQ +QQ + P M YG P+ + + PPPP+P
Sbjct: 585 QQQQQQQQANMNMYPPHMMYGRPHPHQHQHPSMNYFPPPPMP 710
>AW782223
Length = 419
Score = 31.6 bits (70), Expect = 0.56
Identities = 23/84 (27%), Positives = 38/84 (44%)
Frame = +3
Query: 56 PPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADL 115
PPL+ RP P++PR+ PR ++ P++ P + + S+ ++
Sbjct: 36 PPLLLDRP--------PRRPRRLRPRDLRRRSHPPRRVPPSRRAPRPSEVQQ-GLRRRPR 188
Query: 116 LPILLKKNLVQTLPLPRVPNSLPP 139
LP L + LPR+P LPP
Sbjct: 189 LPRLER--------LPRLPRPLPP 236
>CF922465
Length = 538
Score = 31.2 bits (69), Expect = 0.73
Identities = 13/35 (37%), Positives = 20/35 (57%)
Frame = -2
Query: 307 CVFHQGAQGHDVERCFSLKIEVQKLIEDDLIPFEE 341
C H+ + GH +E C + K +VQ I+ + FEE
Sbjct: 444 CACHEESLGHSIEHCMTSKRKVQGPIDAGWLKFEE 340
Score = 30.4 bits (67), Expect = 1.2
Identities = 15/53 (28%), Positives = 26/53 (48%), Gaps = 3/53 (5%)
Frame = -2
Query: 138 PPWYR---PDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIKVL 187
PP++R + C H+ + GH E C K +VQ I+ F++ + +L
Sbjct: 480 PPFFRGYDSNATCACHEESLGHSIEHCMTSKRKVQGPIDAGWLKFEENRV*IL 322
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,853,547
Number of Sequences: 63676
Number of extensions: 309185
Number of successful extensions: 2408
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 2195
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2372
length of query: 348
length of database: 12,639,632
effective HSP length: 98
effective length of query: 250
effective length of database: 6,399,384
effective search space: 1599846000
effective search space used: 1599846000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146784.3


