

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146784.1 - phase: 0
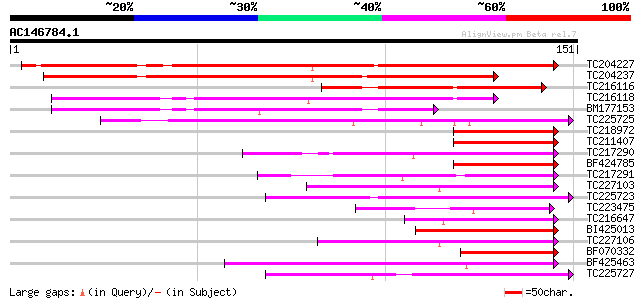
(151 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204227 weakly similar to UP|Q9M6E1 (Q9M6E1) DNA-binding protei... 171 2e-43
TC204237 similar to UP|Q9SST6 (Q9SST6) Elicitor-induced DNA-bind... 148 1e-36
TC216116 similar to UP|Q9M6E1 (Q9M6E1) DNA-binding protein 3, pa... 63 5e-11
TC216118 similar to UP|Q9SST6 (Q9SST6) Elicitor-induced DNA-bind... 57 3e-09
BM177153 similar to GP|17385638|dbj elicitor-induced DNA-binding... 48 2e-06
TC225725 similar to UP|Q9FR29 (Q9FR29) Transcription factor WRKY... 47 5e-06
TC218972 similar to UP|Q9ZPL6 (Q9ZPL6) DNA-binding protein 2, pa... 43 5e-05
TC211407 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding pro... 42 1e-04
TC217290 weakly similar to UP|WR46_ARATH (Q9SKD9) Probable WRKY ... 42 1e-04
BF424785 42 1e-04
TC217291 similar to UP|WR30_ARATH (Q9FL62) Probable WRKY transcr... 42 1e-04
TC227103 similar to UP|WR23_ARATH (O22900) Probable WRKY transcr... 42 1e-04
TC225723 similar to UP|Q9FR29 (Q9FR29) Transcription factor WRKY... 41 2e-04
TC223475 similar to UP|WR13_ARATH (Q9SVB7) Probable WRKY transcr... 40 3e-04
TC216647 similar to UP|WR41_ARATH (Q8H0Y8) Probable WRKY transcr... 40 3e-04
BI425013 40 3e-04
TC227106 similar to UP|WR23_ARATH (O22900) Probable WRKY transcr... 40 6e-04
BF070332 39 7e-04
BF425463 39 7e-04
TC225727 similar to UP|WR40_ARATH (Q9SAH7) Probable WRKY transcr... 39 7e-04
>TC204227 weakly similar to UP|Q9M6E1 (Q9M6E1) DNA-binding protein 3, partial
(25%)
Length = 1110
Score = 171 bits (432), Expect = 2e-43
Identities = 91/149 (61%), Positives = 110/149 (73%), Gaps = 6/149 (4%)
Frame = +1
Query: 4 ENLAHGSGRGKVIDELRRGRELANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTN 63
ENL GS R K I+EL RGR+ A QLK+++N G +DD GS TPFA+ L+KEVL +FTN
Sbjct: 46 ENLG-GSSRRKAIEELLRGRDSAQQLKSVIN--GTYDD--GSATPFAQQLVKEVLMSFTN 210
Query: 64 SLLFLNNNPTSEGSDM------QLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKE 117
SLLFL+NNPTSE + KSEDS ESNCKS++I KE RGCYKRR+ QTWEKE
Sbjct: 211 SLLFLHNNPTSESHHVFNVQVWDSPKSEDSQESNCKSSTI-KEPRGCYKRRRTEQTWEKE 387
Query: 118 SDRPVEDGHQWRKYGQKKILHTDFPRYIY 146
S+ P++DGH WRKYGQK+IL+ FPR Y
Sbjct: 388 SEAPIDDGHHWRKYGQKEILNAKFPRNYY 474
>TC204237 similar to UP|Q9SST6 (Q9SST6) Elicitor-induced DNA-binding protein
homolog, partial (5%)
Length = 477
Score = 148 bits (373), Expect = 1e-36
Identities = 76/127 (59%), Positives = 94/127 (73%), Gaps = 6/127 (4%)
Frame = +3
Query: 10 SGRGKVIDELRRGRELANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLFLN 69
S R K I+EL RGR+ A QL++++N G +D + +T FAE L+KEVL +FTNSL FL
Sbjct: 105 SSRRKAIEELLRGRDCAKQLRSVIN--GSCEDGSSTTPSFAEQLVKEVLMSFTNSLSFLK 278
Query: 70 NNPTSEGSDM------QLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVE 123
NNPTSE D+ + KSEDS ESNCKS SI+KERRGCYKRR+ QTWEKES+ P++
Sbjct: 279 NNPTSESHDVSNVQVCESPKSEDSQESNCKS-SIIKERRGCYKRRRTEQTWEKESEAPID 455
Query: 124 DGHQWRK 130
DGHQWRK
Sbjct: 456 DGHQWRK 476
>TC216116 similar to UP|Q9M6E1 (Q9M6E1) DNA-binding protein 3, partial (26%)
Length = 1525
Score = 63.2 bits (152), Expect = 5e-11
Identities = 33/60 (55%), Positives = 40/60 (66%)
Frame = +2
Query: 84 SEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGHQWRKYGQKKILHTDFPR 143
SEDS ES S K+RRG YKRRK QTW + + +D H WRKYGQK+IL++ FPR
Sbjct: 434 SEDSTESKKGS----KDRRGSYKRRKTEQTWTIVA-QTTDDNHAWRKYGQKEILNSQFPR 598
>TC216118 similar to UP|Q9SST6 (Q9SST6) Elicitor-induced DNA-binding protein
homolog, partial (8%)
Length = 581
Score = 57.4 bits (137), Expect = 3e-09
Identities = 44/136 (32%), Positives = 66/136 (48%), Gaps = 17/136 (12%)
Frame = +1
Query: 12 RGKVIDELRRGRELANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLFLNNN 71
R +VI EL +GR+ A QLK +L + D GS + A++L+ VL +FT +L L ++
Sbjct: 190 RKRVIRELVQGRDYATQLKFLLQKPIGPD---GSVS--AKELVANVLRSFTETLSVLTSS 354
Query: 72 PTSEGSD-----------------MQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTW 114
+ D Q+ + E + +S K+RRG YKRRK QTW
Sbjct: 355 EVAISGDHHRDEVAQNLVISGEDASQVESIDPRSEGSTESKKGSKDRRGSYKRRKTEQTW 534
Query: 115 EKESDRPVEDGHQWRK 130
+ + +D H WRK
Sbjct: 535 TIVA-QTTDDNHAWRK 579
>BM177153 similar to GP|17385638|dbj elicitor-induced DNA-binding protein
homolog {Matricaria chamomilla}, partial (7%)
Length = 429
Score = 48.1 bits (113), Expect = 2e-06
Identities = 42/127 (33%), Positives = 59/127 (46%), Gaps = 24/127 (18%)
Frame = +2
Query: 12 RGKVIDELRRGRELANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTNSL------ 65
R +VI EL +GR+ A QLK +L + D GS + A++L+ VL +F +L
Sbjct: 56 RKRVIRELVQGRDYATQLKFLLQKPIGPD---GSVS--AKELVANVLRSFAETLSVLTSS 220
Query: 66 ------------------LFLNNNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKR 107
L ++ S+ + + SEDS ES S K+RRG YKR
Sbjct: 221 SEDSTSGHDHDDEVIAQNLVISGEDASQVASINDPSSEDSTESRKGS----KDRRGSYKR 388
Query: 108 RKVSQTW 114
RK QTW
Sbjct: 389 RKTEQTW 409
>TC225725 similar to UP|Q9FR29 (Q9FR29) Transcription factor WRKY4, partial
(51%)
Length = 1012
Score = 46.6 bits (109), Expect = 5e-06
Identities = 40/140 (28%), Positives = 61/140 (43%), Gaps = 14/140 (10%)
Frame = +2
Query: 25 LANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLFLNNNPTSEGSDMQLTKS 84
LA +LK + E N T ++ + T N + ++ NP E S + KS
Sbjct: 113 LAEELKRVSAE-------NKKLTEMLTEMCENYNTLRGNLMEYMRKNPDKEHSSSKKRKS 271
Query: 85 EDSLES-----NCKSTSIVKERRGCYKRR-----KVSQTWEKE--SDRP--VEDGHQWRK 130
E + S N S S + C K++ K+S+ + + SD+ V+DG+QWRK
Sbjct: 272 ESNNNSIPMGVNGTSESSSTDEESCKKQKEDIKTKISRVYMRTEASDKSLIVKDGYQWRK 451
Query: 131 YGQKKILHTDFPRYIYFYEF 150
YGQK PR + F
Sbjct: 452 YGQKVTRDNPSPRAYFKCSF 511
>TC218972 similar to UP|Q9ZPL6 (Q9ZPL6) DNA-binding protein 2, partial (33%)
Length = 1177
Score = 43.1 bits (100), Expect = 5e-05
Identities = 15/28 (53%), Positives = 22/28 (78%)
Frame = +2
Query: 119 DRPVEDGHQWRKYGQKKILHTDFPRYIY 146
D+P +DG+ WRKYGQK++ ++FPR Y
Sbjct: 29 DKPADDGYNWRKYGQKQVKGSEFPRSYY 112
Score = 30.8 bits (68), Expect = 0.26
Identities = 11/25 (44%), Positives = 17/25 (68%)
Frame = +2
Query: 122 VEDGHQWRKYGQKKILHTDFPRYIY 146
++DG++WRKYGQK + + R Y
Sbjct: 542 LDDGYRWRKYGQKVVKGNPYXRSYY 616
>TC211407 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding protein,
partial (33%)
Length = 784
Score = 42.0 bits (97), Expect = 1e-04
Identities = 14/28 (50%), Positives = 22/28 (78%)
Frame = +2
Query: 119 DRPVEDGHQWRKYGQKKILHTDFPRYIY 146
D+P +DG+ WRKYGQK++ +++PR Y
Sbjct: 470 DKPADDGYNWRKYGQKQVKGSEYPRSYY 553
>TC217290 weakly similar to UP|WR46_ARATH (Q9SKD9) Probable WRKY
transcription factor 46 (WRKY DNA-binding protein 46),
partial (28%)
Length = 1151
Score = 42.0 bits (97), Expect = 1e-04
Identities = 29/85 (34%), Positives = 43/85 (50%), Gaps = 1/85 (1%)
Frame = +1
Query: 63 NSLLFLNNNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYK-RRKVSQTWEKESDRP 121
N + ++ P + GS KSE LE K+ ++ K+R+ K +V +
Sbjct: 7 NGSMMDSHCPLTNGSP----KSE-VLEPEVKNKNVFKKRKTMSKLTEQVKVRLGTAHEGS 171
Query: 122 VEDGHQWRKYGQKKILHTDFPRYIY 146
++DG+ WRKYGQK IL FPR Y
Sbjct: 172 LDDGYSWRKYGQKDILGAKFPRGYY 246
>BF424785
Length = 412
Score = 42.0 bits (97), Expect = 1e-04
Identities = 14/28 (50%), Positives = 22/28 (78%)
Frame = +2
Query: 119 DRPVEDGHQWRKYGQKKILHTDFPRYIY 146
D+P +DG+ WRKYGQK++ +++PR Y
Sbjct: 254 DKPADDGYNWRKYGQKQVKGSEYPRSYY 337
>TC217291 similar to UP|WR30_ARATH (Q9FL62) Probable WRKY transcription
factor 30 (WRKY DNA-binding protein 30), partial (21%)
Length = 1108
Score = 41.6 bits (96), Expect = 1e-04
Identities = 28/83 (33%), Positives = 39/83 (46%), Gaps = 3/83 (3%)
Frame = +1
Query: 67 FLNNNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRG---CYKRRKVSQTWEKESDRPVE 123
F N +P SE +E K+ + K+R+ C ++ KV E ++
Sbjct: 82 FTNGSPKSE-----------VMEPEIKNKDVFKKRKTMSTCTEQVKVCLGTAHEGS--LD 222
Query: 124 DGHQWRKYGQKKILHTDFPRYIY 146
DG+ WRKYGQK IL FPR Y
Sbjct: 223 DGYSWRKYGQKDILGAKFPRGYY 291
>TC227103 similar to UP|WR23_ARATH (O22900) Probable WRKY transcription
factor 23 (WRKY DNA-binding protein 23), partial (36%)
Length = 980
Score = 41.6 bits (96), Expect = 1e-04
Identities = 22/69 (31%), Positives = 37/69 (52%), Gaps = 2/69 (2%)
Frame = +2
Query: 80 QLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQT--WEKESDRPVEDGHQWRKYGQKKIL 137
Q + +D E K+ +K ++ KR++ + K +EDG++WRKYGQK +
Sbjct: 515 QAEEDDDEEEGQQKTKKQLKPKKTNQKRQREPRFAFMTKSEVDHLEDGYRWRKYGQKAVK 694
Query: 138 HTDFPRYIY 146
++ FPR Y
Sbjct: 695 NSPFPRSYY 721
>TC225723 similar to UP|Q9FR29 (Q9FR29) Transcription factor WRKY4, partial
(54%)
Length = 1636
Score = 40.8 bits (94), Expect = 2e-04
Identities = 25/82 (30%), Positives = 39/82 (47%)
Frame = +1
Query: 69 NNNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGHQW 128
NNN G++ ++S + E +CK +E R +T ++ V+DG+QW
Sbjct: 454 NNNSNLMGTNNGNSESSSTDEESCKKPR--EETIKAKISRVYVRTESSDTSLIVKDGYQW 627
Query: 129 RKYGQKKILHTDFPRYIYFYEF 150
RKYGQK +PR + F
Sbjct: 628 RKYGQKVTRDNPYPRAYFKCSF 693
>TC223475 similar to UP|WR13_ARATH (Q9SVB7) Probable WRKY transcription
factor 13 (WRKY DNA-binding protein 13), partial (16%)
Length = 746
Score = 40.4 bits (93), Expect = 3e-04
Identities = 23/54 (42%), Positives = 31/54 (56%), Gaps = 1/54 (1%)
Frame = +2
Query: 93 KSTSIVKERRGCYKRRKVSQTWEKESDRPV-EDGHQWRKYGQKKILHTDFPRYI 145
K+ V+E R C+K SD V +DG++WRKYGQK + +T PRYI
Sbjct: 533 KARRKVREPRFCFKTM---------SDVDVLDDGYKWRKYGQKVVKNTQHPRYI 667
>TC216647 similar to UP|WR41_ARATH (Q8H0Y8) Probable WRKY transcription
factor 41 (WRKY DNA-binding protein 41), partial (28%)
Length = 1253
Score = 40.4 bits (93), Expect = 3e-04
Identities = 20/49 (40%), Positives = 25/49 (50%), Gaps = 8/49 (16%)
Frame = +1
Query: 106 KRRKVSQTW--------EKESDRPVEDGHQWRKYGQKKILHTDFPRYIY 146
K+RK + W E + P ED + WRKYGQK IL +PR Y
Sbjct: 76 KKRKATPKWMDHVRVSCESGLEGPHEDSYNWRKYGQKDILGAKYPRSYY 222
>BI425013
Length = 281
Score = 40.4 bits (93), Expect = 3e-04
Identities = 18/38 (47%), Positives = 23/38 (60%)
Frame = +2
Query: 109 KVSQTWEKESDRPVEDGHQWRKYGQKKILHTDFPRYIY 146
+V + E + P EDG+ WRKYGQK IL +PR Y
Sbjct: 8 RVRVSCESGLEGPHEDGYNWRKYGQKDILGAKYPRSYY 121
>TC227106 similar to UP|WR23_ARATH (O22900) Probable WRKY transcription
factor 23 (WRKY DNA-binding protein 23), partial (32%)
Length = 783
Score = 39.7 bits (91), Expect = 6e-04
Identities = 21/66 (31%), Positives = 36/66 (53%), Gaps = 2/66 (3%)
Frame = +3
Query: 83 KSEDSLESNCKSTSIVKERRGCYKRRKVSQT--WEKESDRPVEDGHQWRKYGQKKILHTD 140
K+ D + K+ +K ++ KR++ + K +EDG++WRKYGQK + ++
Sbjct: 36 KTVDQAPEHQKTKEQLKAKKTNQKRQREPRFAFMTKSEVDHLEDGYRWRKYGQKAVKNSP 215
Query: 141 FPRYIY 146
FPR Y
Sbjct: 216 FPRSYY 233
>BF070332
Length = 426
Score = 39.3 bits (90), Expect = 7e-04
Identities = 14/26 (53%), Positives = 20/26 (76%)
Frame = +1
Query: 121 PVEDGHQWRKYGQKKILHTDFPRYIY 146
P EDG+ WRKYGQK++ +++PR Y
Sbjct: 316 PSEDGYNWRKYGQKQVKGSEYPRSYY 393
>BF425463
Length = 421
Score = 39.3 bits (90), Expect = 7e-04
Identities = 21/99 (21%), Positives = 46/99 (46%), Gaps = 10/99 (10%)
Frame = +1
Query: 58 LTTFTNSLLFLNNNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKE 117
+++ +N NN+ G ++Q + + + + K++ K++K ++
Sbjct: 61 ISSSSNEATVNNNSEQQSGKELQEEEGGNGGRGDNQDQDKTKKQLKAKKKKKKKNQKKQR 240
Query: 118 SDR----------PVEDGHQWRKYGQKKILHTDFPRYIY 146
R ++DG++WRKYGQK + ++ +PR Y
Sbjct: 241 EPRFAFMTKSEVDHLDDGYKWRKYGQKAVKNSPYPRSYY 357
>TC225727 similar to UP|WR40_ARATH (Q9SAH7) Probable WRKY transcription
factor 40 (WRKY DNA-binding protein 40), partial (41%)
Length = 1177
Score = 39.3 bits (90), Expect = 7e-04
Identities = 26/84 (30%), Positives = 40/84 (46%), Gaps = 2/84 (2%)
Frame = +3
Query: 69 NNNPTSEGSDMQLTKSEDSLESNCKST--SIVKERRGCYKRRKVSQTWEKESDRPVEDGH 126
NNN G++ ++S + E +CK I+K + R +T ++ V+DG+
Sbjct: 48 NNNSNLMGTNNGNSESSSTDEESCKKPREEIIKAKIS----RVYVRTEASDTSLIVKDGY 215
Query: 127 QWRKYGQKKILHTDFPRYIYFYEF 150
QWRKYGQK PR + F
Sbjct: 216 QWRKYGQKVTRDNPCPRAYFKCSF 287
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,456,377
Number of Sequences: 63676
Number of extensions: 77324
Number of successful extensions: 476
Number of sequences better than 10.0: 130
Number of HSP's better than 10.0 without gapping: 463
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 468
length of query: 151
length of database: 12,639,632
effective HSP length: 89
effective length of query: 62
effective length of database: 6,972,468
effective search space: 432293016
effective search space used: 432293016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC146784.1


