

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146777.12 + phase: 0
(238 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
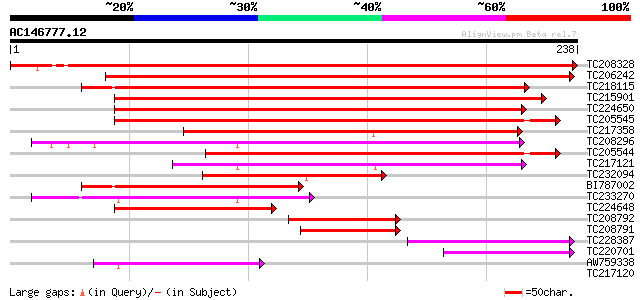
Score E
Sequences producing significant alignments: (bits) Value
TC208328 similar to GB|AAO63325.1|28950803|BT005261 At5g23140 {A... 390 e-109
TC206242 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp prote... 193 7e-50
TC218115 similar to UP|Q94B60 (Q94B60) ATP-dependent Clp proteas... 161 2e-40
TC215901 similar to UP|O82421 (O82421) Clp protease (Fragment) ,... 161 3e-40
TC224650 homologue to UP|O49081 (O49081) Clp-like energy-depende... 148 2e-36
TC205545 similar to PIR|T52454|T52454 ATP-dependent Clp proteina... 140 7e-34
TC217358 homologue to UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp p... 133 6e-32
TC208296 similar to UP|O23548 (O23548) CLP proteinase like prote... 132 1e-31
TC205544 similar to UP|Q93ZD4 (Q93ZD4) At1g12410/F5O11_7, partia... 118 2e-27
TC217121 similar to UP|Q9XJ35 (Q9XJ35) NClpP5 (F2J10.14 protein)... 94 5e-20
TC232094 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp prote... 89 1e-18
BI787002 similar to GP|14596089|gb| ATP-dependent Clp protease-l... 76 1e-14
TC233270 similar to UP|O04018 (O04018) F7G19.1 protein, partial ... 61 5e-10
TC224648 homologue to UP|O49081 (O49081) Clp-like energy-depende... 54 6e-08
TC208792 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp prote... 53 1e-07
TC208791 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp prote... 52 2e-07
TC228387 similar to UP|O04018 (O04018) F7G19.1 protein, partial ... 50 9e-07
TC220701 similar to UP|Q9S834 (Q9S834) ATP-dependent Clp proteas... 49 3e-06
AW759338 similar to GP|16209712|gb| At1g12410/F5O11_7 {Arabidops... 41 4e-04
TC217120 similar to UP|Q9XJ35 (Q9XJ35) NClpP5 (F2J10.14 protein)... 36 0.018
>TC208328 similar to GB|AAO63325.1|28950803|BT005261 At5g23140 {Arabidopsis
thaliana;} , partial (88%)
Length = 1033
Score = 390 bits (1003), Expect = e-109
Identities = 199/243 (81%), Positives = 218/243 (88%), Gaps = 5/243 (2%)
Frame = +2
Query: 1 MRGLFGITKT-----LFNGTKSSFSPCRNRAYSLIPMVIETSSRGERAYDIFSRLLKERI 55
+R + G+ K LF+G K+ + C R+YSLIPMVIE SSRGERAYDIFSRLLKERI
Sbjct: 32 LRAMRGVVKAFAGNLLFHGRKTQ-TQCI-RSYSLIPMVIEHSSRGERAYDIFSRLLKERI 205
Query: 56 VCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPIN 115
+CINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSP+N
Sbjct: 206 ICINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPVN 385
Query: 116 TICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAIHTKQIVRMWDA 175
TIC+GQAASMGSLLL AGAKG+RR+LPNATIMIHQPSGGYSGQAKDIAIHTK IVR+WD+
Sbjct: 386 TICMGQAASMGSLLLAAGAKGERRSLPNATIMIHQPSGGYSGQAKDIAIHTKHIVRVWDS 565
Query: 176 LNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDK 235
LNELY KHTGQ ++VIQ NMDRD FMT +EAKEFG+IDEVIDQRPM LVSDAV +EGKDK
Sbjct: 566 LNELYAKHTGQSVEVIQTNMDRDNFMTPKEAKEFGLIDEVIDQRPMALVSDAVGNEGKDK 745
Query: 236 GSN 238
GSN
Sbjct: 746 GSN 754
>TC206242 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (NClpP1) (ATP-dependent Clp protease proteolytic
subunit ClpP5) , partial (86%)
Length = 1276
Score = 193 bits (490), Expect = 7e-50
Identities = 98/197 (49%), Positives = 136/197 (68%)
Frame = +1
Query: 41 ERAYDIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAG 100
ER + S+L + RI+ G + DD A+++VAQLL+L++ +P+K I MY+NSPGG+VTAG
Sbjct: 427 ERFQSVISQLFQYRIIRCGGAVDDDMANIIVAQLLYLDAVDPNKDIVMYVNSPGGSVTAG 606
Query: 101 LAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAK 160
+AI+DTM++IR ++T+C+G AASMG+ LL AG KG+R +LPN+ IMIHQP GG G
Sbjct: 607 MAIFDTMRHIRPDVSTVCVGLAASMGAFLLSAGTKGKRYSLPNSRIMIHQPLGGAQGGQT 786
Query: 161 DIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRP 220
DI I +++ LN HTGQ LD I ++ DRD+FM+A+EAKE+G+ID VI
Sbjct: 787 DIDIQANEMLHHKANLNGYLAYHTGQSLDKINQDTDRDFFMSAKEAKEYGLIDGVIMNPL 966
Query: 221 MTLVSDAVADEGKDKGS 237
L A EGKD+ S
Sbjct: 967 KALQPLEAAAEGKDRAS 1017
>TC218115 similar to UP|Q94B60 (Q94B60) ATP-dependent Clp protease-like
protein (At5g45390), partial (77%)
Length = 1087
Score = 161 bits (408), Expect = 2e-40
Identities = 79/188 (42%), Positives = 126/188 (67%)
Frame = +3
Query: 31 PMVIETSSRGERAYDIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYL 90
P T++RG D+ LL+ERIV + I D A +++Q+L L++++P+K I +++
Sbjct: 270 PQTPSTAARGAEG-DVMGLLLRERIVFLGSSIDDFVADAIMSQMLLLDAQDPTKDIRLFI 446
Query: 91 NSPGGAVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQ 150
NS GG+++A +AIYD +Q +R+ ++T+ LG AAS S++L G KG+R A+PN IMIHQ
Sbjct: 447 NSTGGSLSATMAIYDAVQLVRADVSTVALGIAASTASVILGGGTKGKRFAMPNTRIMIHQ 626
Query: 151 PSGGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFG 210
P GG SGQA D+ I K+++ + + + TG+ + +QK++DRD +M+ EA E+G
Sbjct: 627 PLGGASGQAIDVEIQAKEVMHNKNNITRIISSFTGRSFEQVQKDIDRDKYMSPIEAVEYG 806
Query: 211 IIDEVIDQ 218
IID VID+
Sbjct: 807 IIDGVIDR 830
>TC215901 similar to UP|O82421 (O82421) Clp protease (Fragment) , partial
(75%)
Length = 1214
Score = 161 bits (407), Expect = 3e-40
Identities = 75/181 (41%), Positives = 124/181 (68%)
Frame = +1
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D + LL++RI+ + + D TA +++QLLFL++E+ K I +++NSPGG+VTAG+ IY
Sbjct: 277 DATNMLLRQRIIFLGSQVDDMTADFIISQLLFLDAEDSKKDIKLFINSPGGSVTAGMGIY 456
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
D M+ ++ ++T+CLG AASMG+ +L +G KG+R +PN+ +MIHQP G G+A +++I
Sbjct: 457 DAMKLCKADVSTVCLGLAASMGAFILASGTKGKRYCMPNSRVMIHQPLGTAGGKATEMSI 636
Query: 165 HTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLV 224
+++ +N++ + TG+P + I+ + DRD FM EAKE+G++D VID LV
Sbjct: 637 RIREMAYHKIKINKILSRITGKPEEQIELDTDRDNFMNPWEAKEYGLVDGVIDDGKPGLV 816
Query: 225 S 225
+
Sbjct: 817 A 819
>TC224650 homologue to UP|O49081 (O49081) Clp-like energy-dependent protease,
partial (72%)
Length = 1067
Score = 148 bits (373), Expect = 2e-36
Identities = 72/173 (41%), Positives = 109/173 (62%)
Frame = +1
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D+ S L + RI+ I P++ A V++QL+ L + N + I +Y+N PGG+ + LAIY
Sbjct: 304 DLTSVLFRNRIIFIGQPVNSQVAQRVISQLVTLATINENADILVYINCPGGSTYSVLAIY 483
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
D M +I+ + T+C G AAS G+LLL G KG R A+PNA IMIHQP G G +D+
Sbjct: 484 DCMSWIKPKVGTVCFGVAASQGALLLAGGEKGMRYAMPNARIMIHQPQSGCGGHVEDVRR 663
Query: 165 HTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVID 217
+ V+ ++++Y TGQPL+ +Q+ +RD F++ EA EFG+ID V++
Sbjct: 664 QVNEAVQSRHKIDKMYSVFTGQPLEKVQQYTERDRFLSVSEALEFGLIDGVLE 822
>TC205545 similar to PIR|T52454|T52454 ATP-dependent Clp proteinase
catalytic chain P2 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (81%)
Length = 1369
Score = 140 bits (352), Expect = 7e-34
Identities = 72/187 (38%), Positives = 116/187 (61%)
Frame = +3
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D+++ L +ER++ I I ++ ++ ++A +L+L+S SK + MY+N PGG +T +AIY
Sbjct: 390 DVWNALYRERVIFIGQEIDEEFSNQILATMLYLDSIENSKKLYMYINGPGGDLTPSMAIY 569
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
DTMQ ++SP+ T C+G A S+ + LL AG K R A+P + + + P+G GQA DI
Sbjct: 570 DTMQSLQSPVATHCVGYAYSLAAFLLAAGEKSNRSAMPLSRVALTSPAGAARGQADDIQN 749
Query: 165 HTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLV 224
+++R+ D L K TGQPL+ I K++ R A+EA E+G+ID ++ RP +
Sbjct: 750 EANELLRIRDYLFNELSKKTGQPLEKITKDLSRMKRFNAQEALEYGLIDRIV--RPPRIK 923
Query: 225 SDAVADE 231
+DA E
Sbjct: 924 ADAPRKE 944
>TC217358 homologue to UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease
proteolytic subunit (Endopeptidase Clp) , partial (76%)
Length = 1519
Score = 133 bits (335), Expect = 6e-32
Identities = 62/143 (43%), Positives = 99/143 (68%), Gaps = 1/143 (0%)
Frame = +1
Query: 74 LLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAG 133
+++L E +K + +++NSPGG V G+AIYDTMQ+++ + T+C+G AASMGS LL G
Sbjct: 19 MVYLSIEEENKDLYLFINSPGGWVIPGIAIYDTMQFVQPDVQTVCMGLAASMGSFLLAGG 198
Query: 134 AKGQRRALPNATIMIHQP-SGGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQ 192
+R A P+A +MIHQP S Y Q + + +++++M + + +YV+ TG+PL VI
Sbjct: 199 EITKRLAFPHARVMIHQPASSFYEAQTGEFILEAEELLKMRETITRVYVQRTGKPLWVIS 378
Query: 193 KNMDRDYFMTAEEAKEFGIIDEV 215
++M+RD FM+A EA+ +GI+D V
Sbjct: 379 EDMERDVFMSAAEAQAYGIVDLV 447
>TC208296 similar to UP|O23548 (O23548) CLP proteinase like protein, partial
(74%)
Length = 1003
Score = 132 bits (332), Expect = 1e-31
Identities = 82/225 (36%), Positives = 126/225 (55%), Gaps = 18/225 (8%)
Frame = +3
Query: 10 TLFNGTK---SSFSPCR----NRAYSLIPMVI---ETSSRGERAYDIFSRLLKERIVCIN 59
+LF+G K SS +P N ++ MVI S+ + D+ S L K RIV +
Sbjct: 9 SLFSGHKLRLSSLNPGSFRGSNAKRGVVTMVIPFQRGSAWEQPPPDLASYLYKNRIVYLG 188
Query: 60 GPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG--------GAVTAGLAIYDTMQYIR 111
+ +++A+ L+L+ E+ KPI +Y+NS G G T AIYD M+Y++
Sbjct: 189 MSLVPSVTELILAEFLYLQYEDDDKPIYLYINSTGTTKGGEKLGYETEAFAIYDVMRYVK 368
Query: 112 SPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAIHTKQIVR 171
PI T+C+G A +LLL AGAKG R ALP++TIMI QP + GQA D+ + +++
Sbjct: 369 PPIFTLCVGNAWGEAALLLAAGAKGNRSALPSSTIMIRQPIARFQGQATDVNLARREVNN 548
Query: 172 MWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVI 216
+ L +LY KH + + I+ ++ R + + EA E+GIID+VI
Sbjct: 549 VKTELVKLYAKHMEKTPEQIEADIQRPKYFSPSEAVEYGIIDKVI 683
>TC205544 similar to UP|Q93ZD4 (Q93ZD4) At1g12410/F5O11_7, partial (53%)
Length = 642
Score = 118 bits (296), Expect = 2e-27
Identities = 61/149 (40%), Positives = 92/149 (60%)
Frame = +2
Query: 83 SKPINMYLNSPGGAVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALP 142
SK + MY+N PGG +T +AIYDTMQ ++SP+ T C+G A S+ + LL AG KG R A+P
Sbjct: 8 SKKLYMYINGPGGDLTPSMAIYDTMQSLQSPVATHCVGYAYSLAAFLLAAGEKGNRSAMP 187
Query: 143 NATIMIHQPSGGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMT 202
+ + + P+G GQA DI +++R+ D L + TGQP++ I K++ R
Sbjct: 188 LSRVALTSPAGAARGQADDIQNEANELLRIRDYLFNELAQKTGQPVEKITKDLSRMKRFN 367
Query: 203 AEEAKEFGIIDEVIDQRPMTLVSDAVADE 231
A+EA E+G+ID ++ RP + +DA E
Sbjct: 368 AQEALEYGLIDRIV--RPPRIKADAPRKE 448
>TC217121 similar to UP|Q9XJ35 (Q9XJ35) NClpP5 (F2J10.14 protein)
(At1g49970/F2J10_5), partial (42%)
Length = 988
Score = 94.0 bits (232), Expect = 5e-20
Identities = 52/159 (32%), Positives = 89/159 (55%), Gaps = 10/159 (6%)
Frame = +2
Query: 69 VVVAQLLFLESENPSKPINMYLNSPG---------GAVTAGLAIYDTMQYIRSPINTICL 119
++VAQ ++L+ +NPSKPI +Y+NS G G+ T +I D M Y+++ + T+
Sbjct: 5 LIVAQFMWLDYDNPSKPIYLYINSSGTLNEKNETVGSETEAYSIADMMSYVKADVYTVNC 184
Query: 120 GQAASMGSLLLCAGAKGQRRALPNATIMIHQPS-GGYSGQAKDIAIHTKQIVRMWDALNE 178
G A ++LL G KG R PN++ ++ P SG D+ I K++ + E
Sbjct: 185 GMAFGQAAMLLSLGTKGYRAVQPNSSTKLYLPKVNRSSGAVIDMWIKAKELEANTEYYIE 364
Query: 179 LYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVID 217
L K TG+ + I K++ R ++ A++A ++GI D++ID
Sbjct: 365 LLAKGTGKSKEEIAKDVQRPKYLQAQDAIDYGIADKIID 481
>TC232094 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (NClpP1) (ATP-dependent Clp protease proteolytic
subunit ClpP5) , partial (49%)
Length = 842
Score = 89.4 bits (220), Expect = 1e-18
Identities = 46/97 (47%), Positives = 62/97 (63%), Gaps = 20/97 (20%)
Frame = +2
Query: 82 PSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPINTICLGQAA------------------ 123
P+K I MY++SPGG+VTAG+AI+DTM++IR ++T+C+G AA
Sbjct: 458 PNKDIVMYVSSPGGSVTAGMAIFDTMRHIRPDVSTVCIGLAASKGAFLLSAGTKGNDSFT 637
Query: 124 --SMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQ 158
S G+ LL AG KG+R +LPN+ IMIHQP G GQ
Sbjct: 638 MSSKGAFLLSAGTKGKRYSLPNSRIMIHQPLSGQGGQ 748
>BI787002 similar to GP|14596089|gb| ATP-dependent Clp protease-like protein
{Arabidopsis thaliana}, partial (31%)
Length = 438
Score = 76.3 bits (186), Expect = 1e-14
Identities = 37/93 (39%), Positives = 62/93 (65%)
Frame = +3
Query: 31 PMVIETSSRGERAYDIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYL 90
P T++RG D+ LL+ERIV + I D A +++QLL L++ +P+K I +++
Sbjct: 162 PQTPSTAARGAEG-DVMVLLLRERIVFLGSSIDDFVADAIMSQLLLLDALDPTKDIRLFI 338
Query: 91 NSPGGAVTAGLAIYDTMQYIRSPINTICLGQAA 123
NS GG+++A +AIYD +Q +R+ ++T+ LG AA
Sbjct: 339 NSTGGSLSATMAIYDAVQLVRADVSTVALGIAA 437
>TC233270 similar to UP|O04018 (O04018) F7G19.1 protein, partial (50%)
Length = 601
Score = 60.8 bits (146), Expect = 5e-10
Identities = 44/140 (31%), Positives = 69/140 (48%), Gaps = 21/140 (15%)
Frame = +1
Query: 10 TLFNGTKSSFSPCRNRAYSLIPMVIETSSRGERAY------------DIFSRLLKERIVC 57
TL N K+S +P + P ++ T ++ ER+ D+ S LL RIV
Sbjct: 184 TLLNTPKNSDTPPYLDIFDS-PKLMATPAQVERSVSYNEHRPRRPPPDLPSLLLHGRIVY 360
Query: 58 INGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG---------GAVTAGLAIYDTMQ 108
I P+ +VVA+L++L+ +P +PI +Y+NS G G T G AIYD M
Sbjct: 361 IGMPLVPAVTELVVAELMYLQYMDPKEPIYIYINSTGTTRDDGETVGMETEGFAIYDAMM 540
Query: 109 YIRSPINTICLGQAASMGSL 128
+++ I+T+ +G A L
Sbjct: 541 QLKNEIHTVAVGSAIGQACL 600
>TC224648 homologue to UP|O49081 (O49081) Clp-like energy-dependent protease,
partial (34%)
Length = 678
Score = 53.9 bits (128), Expect = 6e-08
Identities = 26/68 (38%), Positives = 43/68 (63%)
Frame = +2
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D+ S L + RI+ I P++ A V++QL+ L + + + I +Y+N PGG+ + LAIY
Sbjct: 308 DLSSVLFRNRIIFIGQPVNSQVAQRVISQLVTLATIDENADILVYINCPGGSTYSVLAIY 487
Query: 105 DTMQYIRS 112
D M ++RS
Sbjct: 488 DCMSWVRS 511
>TC208792 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (NClpP1) (ATP-dependent Clp protease proteolytic
subunit ClpP5) , partial (14%)
Length = 739
Score = 53.1 bits (126), Expect = 1e-07
Identities = 26/47 (55%), Positives = 31/47 (65%)
Frame = +3
Query: 118 CLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
C + SMG++LL AG KG R +LPN+ IMIHQP GG G DI I
Sbjct: 597 C*SHSHSMGAILLSAGTKGNRYSLPNSRIMIHQPLGGAQGGQTDIDI 737
>TC208791 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (NClpP1) (ATP-dependent Clp protease proteolytic
subunit ClpP5) , partial (14%)
Length = 618
Score = 52.0 bits (123), Expect = 2e-07
Identities = 25/42 (59%), Positives = 30/42 (70%)
Frame = +2
Query: 123 ASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
+SMG++LL AG KG R +LPN+ IMIHQP GG G DI I
Sbjct: 113 SSMGAILLSAGTKGNRYSLPNSRIMIHQPLGGAQGGQTDIDI 238
>TC228387 similar to UP|O04018 (O04018) F7G19.1 protein, partial (16%)
Length = 800
Score = 50.1 bits (118), Expect = 9e-07
Identities = 24/70 (34%), Positives = 40/70 (56%)
Frame = +3
Query: 168 QIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDA 227
Q++ D L +L KHTG + + M R Y+M A AKEFG+ID ++ + +++D
Sbjct: 162 QVIINRDNLVKLLAKHTGNSEETVANVMKRPYYMDATRAKEFGVIDRILWRGQEKVMADV 341
Query: 228 VADEGKDKGS 237
+ + DKG+
Sbjct: 342 ASPDDWDKGA 371
>TC220701 similar to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (NClpP1) (ATP-dependent Clp protease proteolytic
subunit ClpP5) , partial (22%)
Length = 806
Score = 48.5 bits (114), Expect = 3e-06
Identities = 26/55 (47%), Positives = 33/55 (59%)
Frame = +3
Query: 183 HTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDKGS 237
HTGQ L+ I ++ D D+FM A+EAKE+G ID I L A EGKD+ S
Sbjct: 351 HTGQSLEKINQDTDCDFFMKAKEAKEYGFIDGAIMNPLKALQPLPAAVEGKDRDS 515
>AW759338 similar to GP|16209712|gb| At1g12410/F5O11_7 {Arabidopsis
thaliana}, partial (19%)
Length = 253
Score = 41.2 bits (95), Expect = 4e-04
Identities = 21/73 (28%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Frame = +2
Query: 36 TSSRGERAY-DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG 94
T G R + D+++ L +ER++ I I ++ + ++A +L+L+ + M +N P
Sbjct: 32 TPDEGTRQWADVWNTLHRERVIYIAQEIDEEFTYQILATMLYLDRIQNCTRLYMSINGPD 211
Query: 95 GAVTAGLAIYDTM 107
G + +AIYD M
Sbjct: 212 GDLHTSIAIYDAM 250
>TC217120 similar to UP|Q9XJ35 (Q9XJ35) NClpP5 (F2J10.14 protein)
(At1g49970/F2J10_5), partial (15%)
Length = 713
Score = 35.8 bits (81), Expect = 0.018
Identities = 18/60 (30%), Positives = 36/60 (60%)
Frame = +3
Query: 158 QAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVID 217
+AK++ +T+ + EL K TG+ + I K++ R ++ A++A ++GI D++ID
Sbjct: 36 KAKELEANTEYYI-------ELLAKGTGKSKEEIAKDVQRPKYLQAQDAIDYGIADKIID 194
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,706,712
Number of Sequences: 63676
Number of extensions: 105644
Number of successful extensions: 461
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 455
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 457
length of query: 238
length of database: 12,639,632
effective HSP length: 94
effective length of query: 144
effective length of database: 6,654,088
effective search space: 958188672
effective search space used: 958188672
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146777.12


