

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146777.10 - phase: 1 /pseudo
(406 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
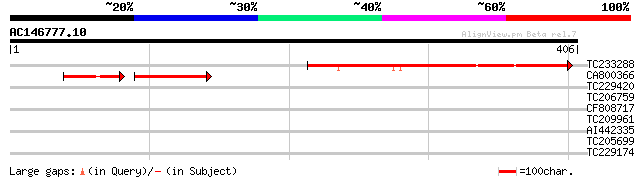
Score E
Sequences producing significant alignments: (bits) Value
TC233288 264 6e-71
CA800366 86 2e-25
TC229420 homologue to GB|AAG30415.1|11118687|AF291666 homeodomai... 33 0.23
TC206759 weakly similar to GB|AAF40464.1|7211993|F13M7 ESTs gb|T... 33 0.30
CF808717 30 2.6
TC209961 similar to UP|ATPL_SULAC (P62020) Membrane-associated A... 29 4.4
AI442335 28 5.7
TC205699 homologue to UP|PHSL_VICFA (P53536) Alpha-1,4 glucan ph... 28 9.8
TC229174 similar to GB|AAL77727.1|18700234|AY078026 AT5g47640/MN... 28 9.8
>TC233288
Length = 835
Score = 264 bits (674), Expect = 6e-71
Identities = 141/216 (65%), Positives = 160/216 (73%), Gaps = 26/216 (12%)
Frame = +3
Query: 214 VVAFYPGVVYSPAYYHHIPGY--LDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGRKM 271
VVAFYPG+VYSPAYY +IPGY +D NPYLITR+DGNVI+AQ WG GGD +ELWNGRK
Sbjct: 3 VVAFYPGIVYSPAYYRYIPGYPKVDTLNPYLITRYDGNVINAQPWGSGGDNRELWNGRKT 182
Query: 272 VD----------EKGSQ-------------VDNSDDVLERRNPLALAHFANHPSKGMLPN 308
+ EKGS+ +++V+ERRNPLALAHFANHP KG+ PN
Sbjct: 183 GEIKPDVKGAEPEKGSERFWKLLSKPLEGYQGGNNEVIERRNPLALAHFANHPPKGVQPN 362
Query: 309 VMICPYDFPLIENDMRAYIPNVLFGNAAEENTERFGSFWFKS-RVPRNNESHVPTTLKTV 367
VMICPYDFPL E++MR YIPNVLFGN AE N RFGSFWFKS RN+ S+VP TLKT+
Sbjct: 363 VMICPYDFPLTESNMRVYIPNVLFGN-AEVNMRRFGSFWFKSGGGSRNSGSNVP-TLKTL 536
Query: 368 VLVATRALQDEELLLNYRLGNTKRCPEWYAPVDEEE 403
VL+ATR LQDEEL LNYRL NTKR PE Y PVDEEE
Sbjct: 537 VLIATRPLQDEELFLNYRLSNTKRRPECYNPVDEEE 644
>CA800366
Length = 395
Score = 85.9 bits (211), Expect(2) = 2e-25
Identities = 41/55 (74%), Positives = 47/55 (84%)
Frame = +1
Query: 90 KNASEADAEEIIEIATKASVADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVND 144
+NA EA AEEIIE+A+KAS ADQQM VQENVH+QIK FCTFMD + LPNEK VN+
Sbjct: 196 ENADEAHAEEIIEMASKASFADQQMQVQENVHSQIKTFCTFMDEILLPNEKMVNE 360
Score = 48.1 bits (113), Expect(2) = 2e-25
Identities = 25/44 (56%), Positives = 32/44 (71%)
Frame = +2
Query: 39 LLQKSWFKCCFQAVKTLAKSPTFAFARDPRQLQYQIDLNRLFLY 82
LL F+ +AVKTLAK P+F +DPRQLQ++ D+NRLFLY
Sbjct: 68 LLMALLFQKFQEAVKTLAKRPSFT--KDPRQLQFEADINRLFLY 193
>TC229420 homologue to GB|AAG30415.1|11118687|AF291666 homeodomain-containing
transcription factor Nkx6.1 {Mus musculus;} , partial
(8%)
Length = 870
Score = 33.1 bits (74), Expect = 0.23
Identities = 18/54 (33%), Positives = 32/54 (58%), Gaps = 3/54 (5%)
Frame = -3
Query: 17 HFSLALCE-PKV*FLYLLCIL*SLLQKSWFKC--CFQAVKTLAKSPTFAFARDP 67
HFSL +C+ P +*F+YL+C + +++WF+ C VK + F+++ P
Sbjct: 205 HFSLIICDRPSI*FVYLICS--TSRRRNWFRSF*CSHPVKCSLTNMHFSWSSPP 50
>TC206759 weakly similar to GB|AAF40464.1|7211993|F13M7 ESTs gb|T76367 and
gb|AA404955 come from this gene. {Arabidopsis thaliana;}
, partial (21%)
Length = 2131
Score = 32.7 bits (73), Expect = 0.30
Identities = 18/59 (30%), Positives = 32/59 (53%), Gaps = 3/59 (5%)
Frame = +3
Query: 156 LPQHSDLRSANGKFVIHQLTILKLKDELGYTLNVKPS---QISHKDAGQGLFLDGVVDV 211
LP + L +A +F+I QL D++ ++V Q+++KD G +F+D +DV
Sbjct: 1218 LPDQALLNTAEWRFLIPQLYKKYPNDDMNLNMSVSSPPDIQVTNKDVGVNIFIDITIDV 1394
>CF808717
Length = 577
Score = 29.6 bits (65), Expect = 2.6
Identities = 13/33 (39%), Positives = 19/33 (57%)
Frame = +1
Query: 44 WFKCCFQAVKTLAKSPTFAFARDPRQLQYQIDL 76
WFKCCF+ V L+K + F R++Q +L
Sbjct: 241 WFKCCFKLVVRLSKRISRPFVIH*REIQQPFNL 339
>TC209961 similar to UP|ATPL_SULAC (P62020) Membrane-associated ATPase C
chain (Sul-ATPase proteolipid chain) , partial (13%)
Length = 586
Score = 28.9 bits (63), Expect = 4.4
Identities = 21/68 (30%), Positives = 29/68 (41%)
Frame = +2
Query: 337 EENTERFGSFWFKSRVPRNNESHVPTTLKTVVLVATRALQDEELLLNYRLGNTKRCPEWY 396
EE + G S P+ E+ +PTTLK R L NYR G + E++
Sbjct: 98 EEEKDEEGDLEECSTTPKGEEAKIPTTLKCPPAPRKR---KPSLKCNYRGGGAR---EFF 259
Query: 397 APVDEEEI 404
P D E +
Sbjct: 260 TPPDLETV 283
>AI442335
Length = 304
Score = 28.5 bits (62), Expect = 5.7
Identities = 11/21 (52%), Positives = 13/21 (61%)
Frame = -2
Query: 328 PNVLFGNAAEENTERFGSFWF 348
PN+ F A E +RFG FWF
Sbjct: 129 PNMKFEEALELTVQRFGMFWF 67
>TC205699 homologue to UP|PHSL_VICFA (P53536) Alpha-1,4 glucan
phosphorylase, L isozyme, chloroplast precursor
(Starch phosphorylase L) , partial (39%)
Length = 1409
Score = 27.7 bits (60), Expect = 9.8
Identities = 8/23 (34%), Positives = 17/23 (73%)
Frame = -2
Query: 3 GYSIRQCLQKLMFYHFSLALCEP 25
G + +C++ ++FY+F++ LC P
Sbjct: 97 GPQLIKCIENIIFYNFTMYLCNP 29
>TC229174 similar to GB|AAL77727.1|18700234|AY078026 AT5g47640/MNJ7_23
{Arabidopsis thaliana;} , partial (63%)
Length = 706
Score = 27.7 bits (60), Expect = 9.8
Identities = 12/29 (41%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Frame = +3
Query: 206 DGVVDVGAVVAFYPGVVY-SPAYYHHIPG 233
D D + +++ G VY SPAY+H +PG
Sbjct: 405 DSSKDSASASSYHQGHVYGSPAYHHQVPG 491
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,761,427
Number of Sequences: 63676
Number of extensions: 264435
Number of successful extensions: 1173
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1163
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1170
length of query: 406
length of database: 12,639,632
effective HSP length: 99
effective length of query: 307
effective length of database: 6,335,708
effective search space: 1945062356
effective search space used: 1945062356
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146777.10


