

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
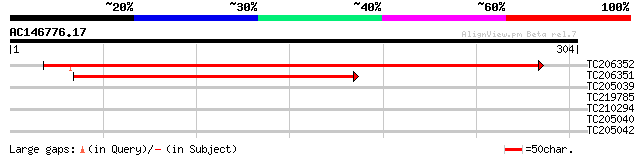
Query= AC146776.17 - phase: 0
(304 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206352 similar to GB|AAN46789.1|24111331|BT001035 At5g18400/F2... 414 e-116
TC206351 weakly similar to GB|AAN46789.1|24111331|BT001035 At5g1... 224 3e-59
TC205039 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding prot... 29 2.3
TC219785 weakly similar to UP|Q8V7F3 (Q8V7F3) ORF1 (Fragment), p... 29 3.0
TC210294 28 6.7
TC205040 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding prot... 27 8.8
TC205042 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding prot... 27 8.8
>TC206352 similar to GB|AAN46789.1|24111331|BT001035 At5g18400/F20L16_120
{Arabidopsis thaliana;} , partial (44%)
Length = 1057
Score = 414 bits (1064), Expect = e-116
Identities = 209/271 (77%), Positives = 236/271 (86%), Gaps = 3/271 (1%)
Frame = +3
Query: 19 SFESFSICYLLLQ---DAAKMYGAVLACTDEAVLPVSQVFDAIRELGNEGVEKLDPLVIT 75
SF+S + L ++ D AK +GAVLACTD AVLPVSQVFDAIRELGNEGVE+ DPLV+T
Sbjct: 15 SFDSVLLLLLPIRIRMDGAKTHGAVLACTDGAVLPVSQVFDAIRELGNEGVEQWDPLVLT 194
Query: 76 SASSLSKFPVESSSVDLVVLIWKSLDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGD 135
SAS LSK PV+SSSVD ++LIW S+D P+DQL E+LRVLK GT LI KSSQSAVGS D
Sbjct: 195 SASLLSKLPVDSSSVDFLILIWLSIDCPVDQLIHEILRVLKVDGTILIRKSSQSAVGSFD 374
Query: 136 KMIPDLENKLLLAGFSEIQALQSSVIKAKKPSWKIGSSFALKKFVKSSPKVQIDFDSDLI 195
K+I LENKLLLAGF+E Q LQS+ IKAKKPSWKIGSSFALKK ++SSPK+QIDFDSDLI
Sbjct: 375 KIISALENKLLLAGFTETQVLQSAGIKAKKPSWKIGSSFALKKVIRSSPKMQIDFDSDLI 554
Query: 196 DKNSLLSEEDLKKPELPSGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNPQSA 255
D++SLL+EEDLKKP+LP GDCEIG TRKACKNC+CGRAEEEEKVLKLGLT EQI+NPQSA
Sbjct: 555 DEDSLLTEEDLKKPQLPPGDCEIGSTRKACKNCTCGRAEEEEKVLKLGLTTEQIDNPQSA 734
Query: 256 CGSCGLGDAFRCSTCPYKGLPAFKMGETVLL 286
CG+CGLGDAFRCSTCPYKGLPAFK+GE V L
Sbjct: 735 CGNCGLGDAFRCSTCPYKGLPAFKLGEKVAL 827
>TC206351 weakly similar to GB|AAN46789.1|24111331|BT001035
At5g18400/F20L16_120 {Arabidopsis thaliana;} , partial
(26%)
Length = 526
Score = 224 bits (572), Expect = 3e-59
Identities = 117/153 (76%), Positives = 132/153 (85%)
Frame = +1
Query: 35 KMYGAVLACTDEAVLPVSQVFDAIRELGNEGVEKLDPLVITSASSLSKFPVESSSVDLVV 94
+M GAVLACTD AVLPVSQVFDAIRELGN G+E+ DPLV+TSAS LSK PV+SSSVD V+
Sbjct: 67 RMDGAVLACTDGAVLPVSQVFDAIRELGNGGLEQWDPLVLTSASLLSKLPVDSSSVDFVI 246
Query: 95 LIWKSLDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQ 154
LIW S+D P+DQL QE+LRVLK GT LI KSSQSAVGS DK+I LENKLLLAGF+E Q
Sbjct: 247 LIWLSIDCPVDQLIQEILRVLKVDGTILIRKSSQSAVGSFDKIISSLENKLLLAGFTEPQ 426
Query: 155 ALQSSVIKAKKPSWKIGSSFALKKFVKSSPKVQ 187
LQS+ IKAKKPSWKIGSSFALKK ++SSPK+Q
Sbjct: 427 VLQSAGIKAKKPSWKIGSSFALKKVIRSSPKMQ 525
>TC205039 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding protein-like,
partial (95%)
Length = 1243
Score = 29.3 bits (64), Expect = 2.3
Identities = 25/105 (23%), Positives = 44/105 (41%), Gaps = 3/105 (2%)
Frame = +3
Query: 159 SVIKAKKPSWKIGSSFAL-KKFVKSSPKVQIDFDSDLIDKNSLLSEED--LKKPELPSGD 215
S++ SW + +F L + ++ K +DL +++E KP SG
Sbjct: 414 SLVAVHSDSWLLSVAFYLGARLNRNERKRLFSLINDLPTVFEVVTERKPVKDKPTADSGS 593
Query: 216 CEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCG 260
G T+++ S G+ + K G E + ++ CGSCG
Sbjct: 594 KSRGSTKRS----SDGQVKSNPKFADEGYEDEDDEHSETLCGSCG 716
>TC219785 weakly similar to UP|Q8V7F3 (Q8V7F3) ORF1 (Fragment), partial (61%)
Length = 913
Score = 28.9 bits (63), Expect = 3.0
Identities = 17/52 (32%), Positives = 25/52 (47%), Gaps = 5/52 (9%)
Frame = +2
Query: 225 CKNCSCGRAEEEEKVLKLGLTAEQINNPQS-----ACGSCGLGDAFRCSTCP 271
C CSC R E +LGL+ E++N+ + CGS + F +T P
Sbjct: 239 CCFCSCIRGNECRPEWQLGLSGERVNSSSTDTGLGCCGSPLTTEHFTSATVP 394
>TC210294
Length = 769
Score = 27.7 bits (60), Expect = 6.7
Identities = 14/36 (38%), Positives = 21/36 (57%), Gaps = 3/36 (8%)
Frame = +3
Query: 222 RKACKNCSCGRAEEEE---KVLKLGLTAEQINNPQS 254
R+ K +C R E+EE + +LGL E++N P S
Sbjct: 195 RRLRKGKACARPEDEETETNIPRLGLLGERLNRPLS 302
>TC205040 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding protein-like,
partial (84%)
Length = 1092
Score = 27.3 bits (59), Expect = 8.8
Identities = 16/53 (30%), Positives = 24/53 (45%)
Frame = +2
Query: 208 KPELPSGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCG 260
KP SG G T+++ S G+ + K G E + ++ CGSCG
Sbjct: 560 KPTADSGSKSRGSTKRS----SDGQVKSNPKFADDGYEDEDDEHSETLCGSCG 706
>TC205042 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding protein-like,
partial (49%)
Length = 676
Score = 27.3 bits (59), Expect = 8.8
Identities = 15/53 (28%), Positives = 24/53 (44%)
Frame = +3
Query: 208 KPELPSGDCEIGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCG 260
KP SG R + K S G+ + K + G ++ + ++ CGSCG
Sbjct: 87 KPTADSGS----KXRGSAKRSSDGQVKSNPKFVDEGYEEDEDEHNETLCGSCG 233
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,370,827
Number of Sequences: 63676
Number of extensions: 205769
Number of successful extensions: 1024
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 1022
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1024
length of query: 304
length of database: 12,639,632
effective HSP length: 97
effective length of query: 207
effective length of database: 6,463,060
effective search space: 1337853420
effective search space used: 1337853420
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146776.17


