

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146776.15 + phase: 0
(536 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
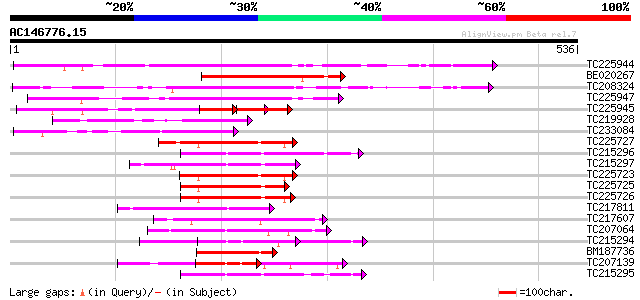
Score E
Sequences producing significant alignments: (bits) Value
TC225944 similar to UP|Q6IET0 (Q6IET0) WRKY transcription factor... 228 4e-60
BE020267 186 2e-47
TC208324 similar to UP|WRK6_ARATH (Q9C519) WRKY transcription fa... 186 2e-47
TC225947 homologue to UP|Q6IET0 (Q6IET0) WRKY transcription fact... 182 3e-46
TC225945 similar to UP|WRK6_ARATH (Q9C519) WRKY transcription fa... 77 3e-30
TC219928 homologue to UP|WR42_ARATH (Q9XEC3) Probable WRKY trans... 122 3e-28
TC233084 similar to UP|WR47_ARATH (Q9ZSI7) Probable WRKY transcr... 115 5e-26
TC225727 similar to UP|WR40_ARATH (Q9SAH7) Probable WRKY transcr... 110 2e-24
TC215296 similar to UP|Q40090 (Q40090) SPF1 protein, partial (26%) 109 3e-24
TC215297 similar to UP|Q40090 (Q40090) SPF1 protein, partial (33%) 106 3e-23
TC225723 similar to UP|Q9FR29 (Q9FR29) Transcription factor WRKY... 105 4e-23
TC225725 similar to UP|Q9FR29 (Q9FR29) Transcription factor WRKY... 103 1e-22
TC225726 similar to UP|Q9SSX8 (Q9SSX8) WIZZ, partial (42%) 103 2e-22
TC217811 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding pro... 102 3e-22
TC217607 similar to UP|Q9SSX8 (Q9SSX8) WIZZ, partial (19%) 101 1e-21
TC207064 similar to UP|Q8W1M6 (Q8W1M6) WRKY-like drought-induced... 100 2e-21
TC215294 similar to UP|Q40090 (Q40090) SPF1 protein, partial (51%) 100 3e-21
BM187736 similar to GP|13506731|gb| WRKY DNA-binding protein 18 ... 99 4e-21
TC207139 similar to UP|Q8W1M6 (Q8W1M6) WRKY-like drought-induced... 99 5e-21
TC215295 similar to UP|Q94IB4 (Q94IB4) WRKY DNA-binding protein ... 99 6e-21
>TC225944 similar to UP|Q6IET0 (Q6IET0) WRKY transcription factor 1, partial
(41%)
Length = 1504
Score = 228 bits (582), Expect = 4e-60
Identities = 165/463 (35%), Positives = 226/463 (48%), Gaps = 5/463 (1%)
Frame = +1
Query: 4 VREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNA--ESDLVSLSLGR 61
+ EN+ K L+ + Y AL+M +++Q ++ +E + E V + G+
Sbjct: 10 MNSENKIRKEMLSHVTGNYTALQMHLVTLMQQNQQRTGSTENEVVQGKVEDKNVGVGGGK 189
Query: 62 VPSNNI---PKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIP 118
VP + P E ++VS + + +E + + ++G+ N
Sbjct: 190 VPRQFLDIGPSGTAEVDDQVS--------DSSSDERTRSSTPQNHNIEAGARDGARNNNG 345
Query: 119 SPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMN 178
E P D + W P+K + D+ + +KARV VRAR + P ++
Sbjct: 346 KSQLGREESP---DSESQGWGPNKLQKVNPSNPMDQSTAEATMRKARVSVRARSEAPMIS 516
Query: 179 DGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLP 238
DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQVQRC +D +IL+TTYEGTHNH LP
Sbjct: 517 DGCQWRKYGQKMAKGNPCPRAYYRCTMAVGCPVRKQVQRCADDRTILVTTYEGTHNHPLP 696
Query: 239 LSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPDGTKSNQLYLSNPA 298
+A AMASTT+AAA+MLLSGS +S G M N NL LP T S
Sbjct: 697 PAAMAMASTTAAAATMLLSGSMSSADGVM-----NPNLLA-RAILPCST-------SMAT 837
Query: 299 LSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPRYPSSTLSFSSPESNPMHW 358
LS+ PT+TLDLT NP+ P F + L P + S ++P + ++
Sbjct: 838 LSASAPFPTVTLDLTHNPNPLQFQRPGAPFQVPF-----LQAQPQNFGSGATPIAQALY- 999
Query: 359 NSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQTYMQKNNNSSNISQHVGLQDS 418
N + S L Q+ S Q Q SQH L D
Sbjct: 1000------------------NQSKFSGLQLS-QDVGSS--QLAPQAPRPPLQPSQHPSLAD- 1113
Query: 419 TISAATKAITADPTFQSALAAALSSLIGNTTNQGNQNQSAGEN 461
T+SAA AIT+DP F + LAAA+SS+IG N N N + N
Sbjct: 1114TVSAAASAITSDPNFTAVLAAAISSIIGGAHNSNNNNNNNSNN 1242
>BE020267
Length = 414
Score = 186 bits (473), Expect = 2e-47
Identities = 97/139 (69%), Positives = 106/139 (75%), Gaps = 3/139 (2%)
Frame = +2
Query: 182 QWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSA 241
QWRKYGQKIAKGNPCPRAYYRCT+AP+CPVRKQVQRC +DMSILITTYEGTHNH +P SA
Sbjct: 2 QWRKYGQKIAKGNPCPRAYYRCTLAPACPVRKQVQRCADDMSILITTYEGTHNHPIPASA 181
Query: 242 TAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNN---LHGLNFYLPDGTKSNQLYLSNPA 298
TAMASTTSAA SMLLSGSSTS A N+ G+NF L D ++NQ+ L P
Sbjct: 182 TAMASTTSAAVSMLLSGSSTSQPTDHSFAYHANSPTLFSGVNFSLLDQPRANQVLLPTP- 358
Query: 299 LSSQHSHPTITLDLTSNPS 317
S H PTITLDLTS PS
Sbjct: 359 --SSHLLPTITLDLTSTPS 409
>TC208324 similar to UP|WRK6_ARATH (Q9C519) WRKY transcription factor 6 (WRKY
DNA-binding protein 6) (AtWRKY6), partial (19%)
Length = 1638
Score = 186 bits (473), Expect = 2e-47
Identities = 152/457 (33%), Positives = 215/457 (46%), Gaps = 2/457 (0%)
Frame = +2
Query: 3 EVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLGRV 62
EV++ENQ L+ LN+I Y AL+ Q ++ + KK L
Sbjct: 353 EVKKENQNLRSMLNEISEHYAALQNQL--LLAMQQKK-------------------LSSS 469
Query: 63 PSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSPVN 122
P NN + N + K AL + +F +N
Sbjct: 470 PRNNEDMQKDSQQNNMEKPALPSCRQF-------------------------------LN 556
Query: 123 SSEVVPIKNDEVVETWPPSKTLNKTMRDAE--DEVAQQTPAKKARVCVRARCDTPTMNDG 180
+ ++ N+ V+ +++A+ +E A + KKARV VRAR ++ M DG
Sbjct: 557 TGKI----NNRVI------------LQEAKIVEEQAFEASCKKARVSVRARSESSLMGDG 688
Query: 181 CQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLS 240
CQWRKYGQKI+KGNPCPRAYYRC + +CPVRKQVQRC ED S++ITTYEG HNHSLP +
Sbjct: 689 CQWRKYGQKISKGNPCPRAYYRCNMGTACPVRKQVQRCAEDESVVITTYEGNHNHSLPPA 868
Query: 241 ATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPDGTKSNQLYLSNPALS 300
A +MASTTSAA M LSGS++S+ GS S +N++L + L+ S S
Sbjct: 869 AKSMASTTSAALKMFLSGSTSSSHGSTYS-YSNSDLF------------SPLFTSTYYPS 1009
Query: 301 SQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPRYPSSTLSFSSPESNPMHWNS 360
+ S PTI LD T TS ++F S ++N+ P +P S +H
Sbjct: 1010ASSSCPTINLDFT------QTSKDNLKFPSVISSNHLQP-FPLS-----------LH--- 1126
Query: 361 FLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQTYMQKNNNSSNISQHVGLQDSTI 420
G+ E I + +++ L D +
Sbjct: 1127--------------------------GQPQQSEGILPS-----------EKNLALVD-VV 1192
Query: 421 SAATKAITADPTFQSALAAALSSLIGNTTNQGNQNQS 457
SAA IT DP+ ++AL AA+SS+IG++ N N +QS
Sbjct: 1193SAA---ITNDPSLKAALEAAVSSIIGDSQNINNHSQS 1294
>TC225947 homologue to UP|Q6IET0 (Q6IET0) WRKY transcription factor 1,
partial (24%)
Length = 793
Score = 182 bits (462), Expect = 3e-46
Identities = 120/302 (39%), Positives = 155/302 (50%), Gaps = 4/302 (1%)
Frame = +2
Query: 18 IMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLGRVPSNN----IPKNDQE 73
+ + Y L+M +++Q+ + +N +E+ G VP +P E
Sbjct: 14 VSSNYANLQMHLAAVLQQQHNQRTENTEQEVVQGKAEERKHGGMVPPRQFLDLVPSGTTE 193
Query: 74 KVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSPVNSSEVVPIKNDE 133
++VS +L ++ STT N + E I +
Sbjct: 194 IDDQVSNSSLGE--------------------RTRSTTPPSCNKNDDKDKKETTDIPHS- 310
Query: 134 VVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDGCQWRKYGQKIAKG 193
K LN T + + + +KARV VRAR + P ++DGCQWRKYGQK+AKG
Sbjct: 311 -------GKLLNHT---TDPSTSPEAAMRKARVSVRARSEAPMISDGCQWRKYGQKMAKG 460
Query: 194 NPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAAS 253
NPCPRAYYRCT+A CPVRKQVQRC ED +IL TTYEGTHNH LP +A AMASTT+AAAS
Sbjct: 461 NPCPRAYYRCTMAVGCPVRKQVQRCAEDRTILTTTYEGTHNHPLPPAAMAMASTTAAAAS 640
Query: 254 MLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPDGTKSNQLYLSNPALSSQHSHPTITLDLT 313
MLLSGS TS G M N NL T++ S LS+ PT+TLDLT
Sbjct: 641 MLLSGSMTSADGIM-----NPNLL---------TRAILPCSSMATLSASAPFPTVTLDLT 778
Query: 314 SN 315
N
Sbjct: 779 HN 784
>TC225945 similar to UP|WRK6_ARATH (Q9C519) WRKY transcription factor 6 (WRKY
DNA-binding protein 6) (AtWRKY6), partial (37%)
Length = 1292
Score = 76.6 bits (187), Expect(2) = 3e-30
Identities = 37/53 (69%), Positives = 44/53 (82%)
Frame = +1
Query: 215 VQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSM 267
VQRC +D +IL+TTYEGTHNH LP +A AMASTT+AAA+MLLSGS +S G M
Sbjct: 1108 VQRCADDRTILVTTYEGTHNHPLPPAAMAMASTTAAAATMLLSGSMSSADGVM 1266
Score = 73.9 bits (180), Expect(2) = 3e-30
Identities = 30/36 (83%), Positives = 33/36 (91%)
Frame = +3
Query: 180 GCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQV 215
G +WRKYGQK+AKGNPCPRAYYRCT+A CPVRKQV
Sbjct: 885 GAKWRKYGQKMAKGNPCPRAYYRCTMAVGCPVRKQV 992
Score = 53.5 bits (127), Expect = 2e-07
Identities = 55/245 (22%), Positives = 101/245 (40%), Gaps = 6/245 (2%)
Frame = +2
Query: 7 ENQRLKMCLNKIMTEYRALEMQFNNMVKQETKK--NNDNNHEEMNAESDLVSLSLGRVPS 64
EN++LK L+ + Y AL+M +++Q ++ + +N + E + G+VP
Sbjct: 380 ENKKLKEMLSHVTGNYTALQMHLVTLMQQNQQRTESTENGVAQGKVEDKNHGVGGGKVPR 559
Query: 65 NNI---PKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSPV 121
+ P E ++VS + + + + + + T +G N +
Sbjct: 560 QFLDIGPSGTAEVDDQVSDSSSDERTRSSTPQ---------DNNTEAGTRDGARN--NNG 706
Query: 122 NSSEVVPIKN-DEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDG 180
N SE+ ++ D + W P+K + D+ + +KARV VRAR + P ++DG
Sbjct: 707 NKSELGREESPDSESQGWGPNKLQKVNPSNPMDQSTAEATMRKARVSVRARSEAPMISDG 886
Query: 181 CQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLS 240
CQ K K G P + + P + Q LI+ Y +N+ P
Sbjct: 887 CQVEKIWTKNG*GESMPSSILQMHYGSWLPSSQTSQ-----FF*LISAYNNNNNNY*PYL 1051
Query: 241 ATAMA 245
+ +A
Sbjct: 1052LSIVA 1066
>TC219928 homologue to UP|WR42_ARATH (Q9XEC3) Probable WRKY transcription
factor 42 (WRKY DNA-binding protein 42), partial (14%)
Length = 518
Score = 122 bits (307), Expect = 3e-28
Identities = 77/189 (40%), Positives = 99/189 (51%)
Frame = +2
Query: 41 NDNNHEEMNAESDLVSLSLGRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLEC 100
N + +E ++ S V S P+NN E +K + A + F++++ G
Sbjct: 50 NADTNETSHSHSSSVIRSQDSPPTNNT-----EVASKKNGGASDEGLVFDQDKKEFGRGI 214
Query: 101 KFETSKSGSTTEGLPNIPSPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTP 160
+ E S S N+P + P +N + E TMR
Sbjct: 215 EREDSPSDQGVAANNNVP------KFSPPRNVDQAEA---------TMR----------- 316
Query: 161 AKKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVE 220
KARV VRAR + P + DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQVQRC E
Sbjct: 317 --KARVSVRARSEAPMITDGCQWRKYGQKMAKGNPCPRAYYRCTMAAGCPVRKQVQRCAE 490
Query: 221 DMSILITTY 229
D +ILITTY
Sbjct: 491 DRTILITTY 517
>TC233084 similar to UP|WR47_ARATH (Q9ZSI7) Probable WRKY transcription
factor 47 (WRKY DNA-binding protein 47), partial (13%)
Length = 614
Score = 115 bits (288), Expect = 5e-26
Identities = 78/218 (35%), Positives = 106/218 (47%), Gaps = 5/218 (2%)
Frame = +3
Query: 4 VREENQRLKMCLNKIMTEYRALEMQF-----NNMVKQETKKNNDNNHEEMNAESDLVSLS 58
++EEN +L+ L+ I Y L+ Q N ETK + +++ +D
Sbjct: 36 LQEENNKLRNVLDHITKSYTQLQAQLFIALQNLPQNMETKIVDPGTSRKLDVVNDA---- 203
Query: 59 LGRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIP 118
S + K DQ+ VS NN E +K L N+
Sbjct: 204 -----SVSDEKTDQD----VSVSRSNNAEVMSKTH-----------DHDDPQLTKLLNLG 323
Query: 119 SPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMN 178
+ + ++W SK L + + AE A+Q P +KARV VRAR + P ++
Sbjct: 324 KQACPDAAEDVLDRSSSQSWGSSK-LEEQPKTAEQLPAEQIPLRKARVSVRARSEAPMIS 500
Query: 179 DGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQ 216
DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQVQ
Sbjct: 501 DGCQWRKYGQKMAKGNPCPRAYYRCTMAVGCPVRKQVQ 614
>TC225727 similar to UP|WR40_ARATH (Q9SAH7) Probable WRKY transcription
factor 40 (WRKY DNA-binding protein 40), partial (41%)
Length = 1177
Score = 110 bits (274), Expect = 2e-24
Identities = 58/136 (42%), Positives = 85/136 (61%), Gaps = 4/136 (2%)
Frame = +3
Query: 141 SKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTM--NDGCQWRKYGQKIAKGNPCPR 198
S T ++ + +E+ + AK +RV VR ++ DG QWRKYGQK+ + NPCPR
Sbjct: 96 SSTDEESCKKPREEIIK---AKISRVYVRTEASDTSLIVKDGYQWRKYGQKVTRDNPCPR 266
Query: 199 AYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSG 258
AY++C+ APSCPV+K+VQR V+D S+L+ TYEG HNH P ++ M +T+ + S+ L
Sbjct: 267 AYFKCSFAPSCPVKKKVQRSVDDQSVLVATYEGEHNH--PQFSSQMEATSGSGRSVTLGS 440
Query: 259 --SSTSNSGSMPSAQT 272
+ S S S P+ T
Sbjct: 441 VPCTASLSTSTPTLVT 488
>TC215296 similar to UP|Q40090 (Q40090) SPF1 protein, partial (26%)
Length = 1278
Score = 109 bits (273), Expect = 3e-24
Identities = 64/173 (36%), Positives = 95/173 (53%)
Frame = +2
Query: 162 KKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVED 221
++ RV V+ D ++DG +WRKYGQK+ KGNP PR+YY+CT P CPVRK V+R +D
Sbjct: 287 REPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTF-PGCPVRKHVERASQD 463
Query: 222 MSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNF 281
+ +ITTYEG HNH +P A + S + S+ + ++T+N+ S+ ++ + NN +
Sbjct: 464 LRAVITTYEGKHNHDVP--AARGSGNNSISRSLPIITNTTNNTTSVATSISTNNNSLQSL 637
Query: 282 YLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNN 334
P + L NP + QHS + NP S + SYNN
Sbjct: 638 RPPAPPERPSLSHFNP--NMQHSSGSFGFSGFGNPLMGSYMN-----QQSYNN 775
>TC215297 similar to UP|Q40090 (Q40090) SPF1 protein, partial (33%)
Length = 1298
Score = 106 bits (264), Expect = 3e-23
Identities = 61/172 (35%), Positives = 94/172 (54%), Gaps = 10/172 (5%)
Frame = +2
Query: 114 LPNIPSPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDA-------EDE---VAQQTPAKK 163
+ ++ +P NSS + +++D+ T + DA E+E + ++
Sbjct: 191 MDSVATPENSS--ISMEDDDFDHTKSGGDEFDNDEPDAKRWRIEGENEGMPAIESRTVRE 364
Query: 164 ARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMS 223
RV + D ++DG +WRKYGQK+ KGNP PR+YY+CT P CPVRK V+R +D+
Sbjct: 365 PRVVFQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTF-PGCPVRKHVERASQDLR 541
Query: 224 ILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNN 275
+ITTYEG HNH +P A S ++ L ++T+N+ + S TNNN
Sbjct: 542 AVITTYEGKHNHDVP---AARGSGNNSMNRSLPITNTTNNTSAATSLYTNNN 688
Score = 37.7 bits (86), Expect = 0.013
Identities = 20/50 (40%), Positives = 30/50 (60%)
Frame = +2
Query: 203 CTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAA 252
CT P+CP +K+V+R + D I Y+GTHNH P +A + + S+ A
Sbjct: 2 CTY-PNCPTKKKVERSL-DGQITEIVYKGTHNHPKPQAAKRNSLSASSLA 145
>TC225723 similar to UP|Q9FR29 (Q9FR29) Transcription factor WRKY4, partial
(54%)
Length = 1636
Score = 105 bits (263), Expect = 4e-23
Identities = 55/116 (47%), Positives = 76/116 (65%), Gaps = 4/116 (3%)
Frame = +1
Query: 161 AKKARVCVRARCDTPTM--NDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRC 218
AK +RV VR ++ DG QWRKYGQK+ + NP PRAY++C+ APSCPV+K+VQR
Sbjct: 553 AKISRVYVRTESSDTSLIVKDGYQWRKYGQKVTRDNPYPRAYFKCSFAPSCPVKKKVQRS 732
Query: 219 VEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSG--SSTSNSGSMPSAQT 272
V+D S+L+ TYEG HNH P +++ M +T+ + S+ L S S S S P+ T
Sbjct: 733 VDDHSVLLATYEGEHNH--PQASSQMEATSGSGRSVTLGSVPCSASLSTSTPTLVT 894
>TC225725 similar to UP|Q9FR29 (Q9FR29) Transcription factor WRKY4, partial
(51%)
Length = 1012
Score = 103 bits (258), Expect = 1e-22
Identities = 50/105 (47%), Positives = 71/105 (67%), Gaps = 2/105 (1%)
Frame = +2
Query: 162 KKARVCVRARCDTPTM--NDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCV 219
K +RV +R ++ DG QWRKYGQK+ + NP PRAY++C+ APSCPV+K+VQR V
Sbjct: 374 KISRVYMRTEASDKSLIVKDGYQWRKYGQKVTRDNPSPRAYFKCSFAPSCPVKKKVQRSV 553
Query: 220 EDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNS 264
+D S+L+ TYEG HNH+ P + M TT + S+ S S +S++
Sbjct: 554 DDQSVLVATYEGEHNHTHP---SQMEVTTGSNRSVSCSASLSSSA 679
>TC225726 similar to UP|Q9SSX8 (Q9SSX8) WIZZ, partial (42%)
Length = 909
Score = 103 bits (257), Expect = 2e-22
Identities = 54/113 (47%), Positives = 71/113 (62%), Gaps = 4/113 (3%)
Frame = +1
Query: 162 KKARVCVRARCDTPTM--NDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCV 219
K +RV +R ++ DG QWRKYGQK+ + NP PRAY++C+ APSCPV+K+VQR V
Sbjct: 61 KISRVYMRTEASDKSLIVKDGYQWRKYGQKVTRDNPSPRAYFKCSFAPSCPVKKKVQRSV 240
Query: 220 EDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSG--SSTSNSGSMPSA 270
+D S+L+ TYEG HNH P + M TT + M L S S S S P+A
Sbjct: 241 DDQSVLVATYEGEHNHPHP---SQMEVTTGSNRCMTLGSVPCSASLSSSPPTA 390
>TC217811 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding protein,
partial (28%)
Length = 1010
Score = 102 bits (255), Expect = 3e-22
Identities = 54/148 (36%), Positives = 79/148 (52%)
Frame = +2
Query: 103 ETSKSGSTTEGLPNIPSPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAK 162
+TS G+ L +P S EV + N E + P+ T + Q
Sbjct: 158 QTSNQGAPPRQL--LPGSNESEEVGNVDNREEADDGEPNPKRRNTDVGVSEVPLSQKTVT 331
Query: 163 KARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDM 222
+ ++ V+ R + ++DG +WRKYGQK+ KGNP PR+YY+CT A C VRK V+R D
Sbjct: 332 EPKIIVQTRSEVDLLDDGYRWRKYGQKVVKGNPHPRSYYKCTSA-GCNVRKHVERASMDP 508
Query: 223 SILITTYEGTHNHSLPLSATAMASTTSA 250
+ITTYEG HNH +P + + +T S+
Sbjct: 509 KAVITTYEGKHNHDVPAARNSSHNTASS 592
>TC217607 similar to UP|Q9SSX8 (Q9SSX8) WIZZ, partial (19%)
Length = 1236
Score = 101 bits (251), Expect = 1e-21
Identities = 67/187 (35%), Positives = 91/187 (47%), Gaps = 23/187 (12%)
Frame = +1
Query: 137 TWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRA--RCDTPTMNDGCQWRKYGQKIAKGN 194
T PP NK R + T K ++ VR + D+ + DG QWRKYGQK+ K N
Sbjct: 355 TVPPMFDTNKRPR-----LELPTAKKPLQIFVRTHPKDDSLIVKDGYQWRKYGQKVTKDN 519
Query: 195 PCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNH------------------- 235
PRAY+RC++AP CPV+K+VQRC+ D SI++ TY+G HNH
Sbjct: 520 ASPRAYFRCSMAPMCPVKKKVQRCLHDKSIVVATYDGEHNHAAIHESSSSTSKGSSPVVN 699
Query: 236 SLPLSATAMASTTSAAASMLLSG-SSTSNSGSMPSA-QTNNNLHGLNFYLPDGTKSNQLY 293
+LPL + + + LSG S T + A Q NNN G N + + S
Sbjct: 700 NLPLMTSILNDKEPMNIGLALSGWSQTDHRRHCEDAMQQNNNNGGSNIRIEEYVSS---L 870
Query: 294 LSNPALS 300
+ NP S
Sbjct: 871 IKNPDFS 891
>TC207064 similar to UP|Q8W1M6 (Q8W1M6) WRKY-like drought-induced protein,
partial (42%)
Length = 1782
Score = 100 bits (248), Expect = 2e-21
Identities = 63/183 (34%), Positives = 91/183 (49%), Gaps = 9/183 (4%)
Frame = +3
Query: 131 NDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDGCQWRKYGQKI 190
NDEV + P SK + A D P ++ RV V+ + ++DG +WRKYGQK+
Sbjct: 48 NDEVDDDDPFSKRRKMDVGIA-DITPVVKPIREPRVVVQTLSEVDILDDGYRWRKYGQKV 224
Query: 191 AKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSATA---MAST 247
+GNP PR+YY+CT CPVRK V+R D +ITTYEG HNH +P + + MA
Sbjct: 225 VRGNPNPRSYYKCT-NTGCPVRKHVERASHDPKAVITTYEGKHNHDVPTARNSCHDMAGP 401
Query: 248 TSAAASMLLSGSSTS------NSGSMPSAQTNNNLHGLNFYLPDGTKSNQLYLSNPALSS 301
SA+ + + G P+A+ +N G G +Q++ SN
Sbjct: 402 ASASGQTRVRPEESDTISLDLGMGISPAAENTSNSQGRMMLSEFG--DSQIHTSNSNFKF 575
Query: 302 QHS 304
H+
Sbjct: 576 VHT 584
>TC215294 similar to UP|Q40090 (Q40090) SPF1 protein, partial (51%)
Length = 1737
Score = 99.8 bits (247), Expect = 3e-21
Identities = 68/217 (31%), Positives = 98/217 (44%), Gaps = 1/217 (0%)
Frame = +1
Query: 123 SSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDGCQ 182
SS+ DE E P +K + ++ RV V+ D ++DG +
Sbjct: 757 SSQKCKSGGDEYDEDEPDAKRWKIEGENEGMSAPGSRTVREPRVVVQTTSDIDILDDGYR 936
Query: 183 WRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSAT 242
WRKYGQK+ KGNP PR+YY+CT P CPVRK V+R D+ +ITTYEG HNH +P +
Sbjct: 937 WRKYGQKVVKGNPNPRSYYKCT-HPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARG 1113
Query: 243 AMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNF-YLPDGTKSNQLYLSNPALSS 301
SGS + N +A + N + LP +S+ + + +
Sbjct: 1114--------------SGSHSVNRPMPNNASNHTNTAATSVRLLPVIHQSDNSLQNQRSQAP 1251
Query: 302 QHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQL 338
TL++ +P + S F SY N QL
Sbjct: 1252PEGQSPFTLEMLQSPGSFGFSG-FGNPMQSYVNQQQL 1359
Score = 75.5 bits (184), Expect = 6e-14
Identities = 43/107 (40%), Positives = 59/107 (54%), Gaps = 9/107 (8%)
Frame = +1
Query: 178 NDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSL 237
+DG WRKYGQK KG+ PR+YY+CT P+CP +K+V+R + D I Y+GTHNH
Sbjct: 406 DDGYNWRKYGQKQVKGSENPRSYYKCTY-PNCPTKKKVERSL-DGQITEIVYKGTHNHPK 579
Query: 238 PLSATAMASTTSAAA---------SMLLSGSSTSNSGSMPSAQTNNN 275
P + +S +S+ A + +T SG M SA T N
Sbjct: 580 PQNTRRNSSNSSSLAIPHSNSIRTEIPDQSYATHGSGQMDSAATPEN 720
>BM187736 similar to GP|13506731|gb| WRKY DNA-binding protein 18 {Arabidopsis
thaliana}, partial (26%)
Length = 437
Score = 99.4 bits (246), Expect = 4e-21
Identities = 45/77 (58%), Positives = 57/77 (73%)
Frame = +3
Query: 177 MNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHS 236
+ DG QWRKYGQK+ + NP PRAY++C+ APSCPV+K+VQR VED S+L+TTYEG HNH
Sbjct: 123 VRDGYQWRKYGQKVTRDNPSPRAYFKCSYAPSCPVKKKVQRSVEDPSVLVTTYEGEHNHG 302
Query: 237 LPLSATAMASTTSAAAS 253
TA S S++ S
Sbjct: 303 -QQHQTAEISINSSSKS 350
>TC207139 similar to UP|Q8W1M6 (Q8W1M6) WRKY-like drought-induced protein,
partial (85%)
Length = 1774
Score = 99.0 bits (245), Expect = 5e-21
Identities = 71/229 (31%), Positives = 104/229 (45%), Gaps = 12/229 (5%)
Frame = +2
Query: 103 ETSKSGSTTEGLPNIPSPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAK 162
E+S + +GL + V++ + ND+ P K D P +
Sbjct: 761 ESSPVATNDDGLEGVAGFVSNRTNEEVDNDD------PFSKRRKMELGNVDITPVVKPIR 922
Query: 163 KARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDM 222
+ RV V+ + ++DG +WRKYGQK+ +GNP PR+YY+CT A CPVRK V+R D
Sbjct: 923 EPRVVVQTLSEVDILDDGYRWRKYGQKVVRGNPNPRSYYKCTNA-GCPVRKHVERASHDP 1099
Query: 223 SILITTYEGTHNHSLPL---SATAMASTTSAAASMLLSGSSTSNS-------GSMPSAQT 272
+ITTYEG HNH +P S+ MA +AA + S++ G +A+
Sbjct: 1100KAVITTYEGKHNHDVPAARNSSHDMAVPAAAAGGQTRTKLEESDTISLDLGMGISSAAEH 1279
Query: 273 NNNLHGLNFYLPDGTKSNQLYLSNPALSSQHSHPTI--TLDLTSNPSNS 319
+N G + G SN S P L+ +SNP S
Sbjct: 1280RSNGQGKMLHSEFGDTQTHTSSSNFKFVHTTSAPVYFGVLNNSSNPFGS 1426
Score = 65.9 bits (159), Expect = 4e-11
Identities = 31/63 (49%), Positives = 41/63 (64%)
Frame = +2
Query: 176 TMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNH 235
T +DG WRKYGQK+ KG+ PR+YY+CT P+C V+K +R D I Y+GTH+H
Sbjct: 434 TSDDGYNWRKYGQKLVKGSEFPRSYYKCT-HPNCEVKKLFERS-HDGQITEIVYKGTHDH 607
Query: 236 SLP 238
P
Sbjct: 608 PKP 616
>TC215295 similar to UP|Q94IB4 (Q94IB4) WRKY DNA-binding protein (Fragment),
partial (34%)
Length = 925
Score = 98.6 bits (244), Expect = 6e-21
Identities = 61/176 (34%), Positives = 88/176 (49%)
Frame = +1
Query: 162 KKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVED 221
++ RV V+ D ++DG +WRKYGQK+ KGNP PR+YY+CT P CPVRK V+R D
Sbjct: 16 REPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCT-HPGCPVRKHVERASHD 192
Query: 222 MSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNF 281
+ +ITTYEG HNH +P A + + S M + S+ +N+ + + H
Sbjct: 193 LRAVITTYEGKHNHDVP--AARGSGSHSVNRPMPNNASNPTNTAATAISPLQVIQHS--- 357
Query: 282 YLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQ 337
D + NQ + P TL++ +P + S F SY N Q
Sbjct: 358 ---DNSHQNQRSQAPP-----EGQSPFTLEMLQSPGSFGFSG-FGNPMQSYMNQQQ 498
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.306 0.121 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,921,597
Number of Sequences: 63676
Number of extensions: 340306
Number of successful extensions: 2730
Number of sequences better than 10.0: 215
Number of HSP's better than 10.0 without gapping: 2506
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2639
length of query: 536
length of database: 12,639,632
effective HSP length: 102
effective length of query: 434
effective length of database: 6,144,680
effective search space: 2666791120
effective search space used: 2666791120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146776.15


