

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146775.17 + phase: 0 /pseudo
(140 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
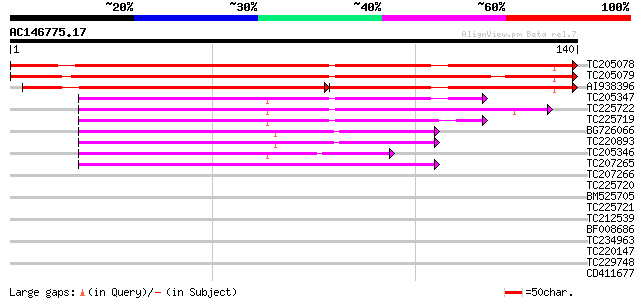
Score E
Sequences producing significant alignments: (bits) Value
TC205078 weakly similar to UP|Q944L1 (Q944L1) At1g70780/F5A18_4,... 189 5e-49
TC205079 weakly similar to UP|Q944L1 (Q944L1) At1g70780/F5A18_4,... 187 2e-48
AI938396 124 4e-45
TC205347 53 5e-08
TC225722 49 6e-07
TC225719 48 1e-06
BG726066 48 2e-06
TC220893 48 2e-06
TC205346 47 4e-06
TC207265 similar to UP|O49657 (O49657) Predicted protein, partia... 40 5e-04
TC207266 weakly similar to UP|O49657 (O49657) Predicted protein,... 39 0.001
TC225720 35 0.009
BM525705 weakly similar to GP|2827553|emb| predicted protein {Ar... 34 0.020
TC225721 homologue to UP|Q7S1C8 (Q7S1C8) Predicted protein, part... 34 0.026
TC212539 33 0.057
BF008686 32 0.13
TC234963 homologue to UP|Q9FH57 (Q9FH57) GATA-binding transcript... 29 0.83
TC220147 28 1.8
TC229748 27 3.1
CD411677 27 4.1
>TC205078 weakly similar to UP|Q944L1 (Q944L1) At1g70780/F5A18_4, partial
(66%)
Length = 911
Score = 189 bits (479), Expect = 5e-49
Identities = 102/142 (71%), Positives = 113/142 (78%), Gaps = 2/142 (1%)
Frame = +2
Query: 1 MLLSKQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLP 60
+ +SKQKKNQNA +RLLISINVLGSAGPIRFVVNEEELV AVIDT LKSYAREGRLP
Sbjct: 275 LTMSKQKKNQNA----KRLLISINVLGSAGPIRFVVNEEELVAAVIDTALKSYAREGRLP 442
Query: 61 VLGNDHSAFFLYCPHLGSDAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSL 120
VLGN+ + F LYCPHLGSD ALSPW++IGSHG RNFVL KKP+ A DGS S
Sbjct: 443 VLGNNTTGFVLYCPHLGSD-ALSPWERIGSHGVRNFVLFKKPEG----VADVDGSTALSR 607
Query: 121 PRRGSGSWKSWFN--LNLKVSS 140
RGSGSWK+WFN LNLK+S+
Sbjct: 608 SSRGSGSWKAWFNKSLNLKIST 673
>TC205079 weakly similar to UP|Q944L1 (Q944L1) At1g70780/F5A18_4, partial
(64%)
Length = 996
Score = 187 bits (474), Expect = 2e-48
Identities = 100/142 (70%), Positives = 112/142 (78%), Gaps = 2/142 (1%)
Frame = +3
Query: 1 MLLSKQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLP 60
ML+SKQKK+Q N +RLLISINVLGSAGPIRFVVNEEELV AVIDT LKSYAREGRLP
Sbjct: 345 MLMSKQKKSQIQN--AKRLLISINVLGSAGPIRFVVNEEELVAAVIDTALKSYAREGRLP 518
Query: 61 VLGNDHSAFFLYCPHLGSDAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSL 120
VLGN+ + F LYCPHLGSD ALSPWD+IGSHG RNF+LCKKP+ + G SS
Sbjct: 519 VLGNNTTGFALYCPHLGSD-ALSPWDRIGSHGVRNFMLCKKPEGVADVDGDASGISRSS- 692
Query: 121 PRRGSGSWKSWFN--LNLKVSS 140
RG GSWK+WFN LNLK+S+
Sbjct: 693 --RGIGSWKAWFNKSLNLKIST 752
>AI938396
Length = 456
Score = 124 bits (312), Expect(2) = 4e-45
Identities = 65/76 (85%), Positives = 67/76 (87%)
Frame = +2
Query: 4 SKQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLG 63
SKQKKNQNAN RLLISINVLGSAGPIRFVVNEEELV AVIDT LKSYAREGRLPVLG
Sbjct: 2 SKQKKNQNAN----RLLISINVLGSAGPIRFVVNEEELVAAVIDTALKSYAREGRLPVLG 169
Query: 64 NDHSAFFLYCPHLGSD 79
N+ + F LYCPHLGSD
Sbjct: 170NNTTGFVLYCPHLGSD 217
Score = 72.8 bits (177), Expect(2) = 4e-45
Identities = 37/63 (58%), Positives = 44/63 (69%), Gaps = 2/63 (3%)
Frame = +1
Query: 80 AALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSLPRRGSGSWKSWFN--LNLK 137
+ LSPW++IGSHG RNFVL KKP+ A DGS S RGSGSWK+WFN LNLK
Sbjct: 229 STLSPWERIGSHGVRNFVLFKKPEG----VADVDGSTALSRSSRGSGSWKAWFNKSLNLK 396
Query: 138 VSS 140
+S+
Sbjct: 397 IST 405
>TC205347
Length = 1514
Score = 52.8 bits (125), Expect = 5e-08
Identities = 31/102 (30%), Positives = 53/102 (51%), Gaps = 1/102 (0%)
Frame = +2
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVL-GNDHSAFFLYCPHL 76
+LL+ + +GS GP++ V+ E V ++ ++ Y +EGR P+L ND S F L+
Sbjct: 572 KLLLKVTSMGSLGPVQVVMTPESTVGDLVAAAVRQYVKEGRRPILPSNDPSHFDLHYSQF 751
Query: 77 GSDAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTS 118
+ +L +K+ G+RNF +C + DG GT+
Sbjct: 752 SLE-SLDREEKLRDIGSRNFFMCPRKCG----GGGGDGRGTT 862
>TC225722
Length = 1140
Score = 49.3 bits (116), Expect = 6e-07
Identities = 32/125 (25%), Positives = 59/125 (46%), Gaps = 8/125 (6%)
Frame = +2
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVL-GNDHSAFFLYCPHL 76
++L+ + +L S P++ ++ E V +I ++ Y +EGR P+L + S F L+
Sbjct: 383 KMLLKVTMLRSLAPVQMLMTPESTVRDLIAAAVRQYVKEGRRPILPAIEASDFDLHYSQF 562
Query: 77 GSDAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSLPRR-------GSGSWK 129
+ +L +K+ G+RNF LC + A T E + + + + G G
Sbjct: 563 SLE-SLDRKEKLLELGSRNFFLCPRKAATTVEGSVTTSFASCAKEAKKERQGCGGGGGGF 739
Query: 130 SWFNL 134
+WF L
Sbjct: 740 AWFKL 754
>TC225719
Length = 1083
Score = 48.1 bits (113), Expect = 1e-06
Identities = 31/102 (30%), Positives = 53/102 (51%), Gaps = 1/102 (0%)
Frame = +1
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVL-GNDHSAFFLYCPHL 76
++L+ + VL S P++ ++ V +I ++ Y +EGR P+L ++ S F L+
Sbjct: 460 KMLLKVTVLRSLAPVQVLMTPGSTVRDLIAAAVRQYVKEGRRPILPSSEVSEFDLHYSQF 639
Query: 77 GSDAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTS 118
+ +L +K+ G+RNF LC + A T EDG TS
Sbjct: 640 SLE-SLDRKEKLLELGSRNFFLCPRKPATT----VEDGVTTS 750
>BG726066
Length = 419
Score = 47.8 bits (112), Expect = 2e-06
Identities = 29/90 (32%), Positives = 49/90 (54%), Gaps = 1/90 (1%)
Frame = +3
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGN-DHSAFFLYCPHL 76
+LL+++ + G ++ +++ E+ V +I L Y +E R P L N D + L+
Sbjct: 90 KLLLNVTIESCLGAVQVLLSPEDTVADLIKAALAFYEKEKRRPFLKNTDPKCYDLHFSSF 269
Query: 77 GSDAALSPWDKIGSHGARNFVLCKKPQAAT 106
++ L DK+ S G+RNF LC KP +AT
Sbjct: 270 TLES-LKADDKLVSLGSRNFFLCSKPPSAT 356
>TC220893
Length = 918
Score = 47.8 bits (112), Expect = 2e-06
Identities = 30/90 (33%), Positives = 49/90 (54%), Gaps = 1/90 (1%)
Frame = +1
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGN-DHSAFFLYCPHL 76
+LL+++ + G ++ +++ E+ V +I L Y RE R P L N D + L+
Sbjct: 388 KLLLNVTIENCLGAVQVLLSPEDTVADLIKAALAFYEREKRRPFLKNTDPKCYDLHFSSF 567
Query: 77 GSDAALSPWDKIGSHGARNFVLCKKPQAAT 106
++ L P DK+ + G+RNF LC K AAT
Sbjct: 568 TLES-LKPDDKLVNLGSRNFFLCCKAPAAT 654
>TC205346
Length = 780
Score = 46.6 bits (109), Expect = 4e-06
Identities = 27/79 (34%), Positives = 45/79 (56%), Gaps = 1/79 (1%)
Frame = +2
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVL-GNDHSAFFLYCPHL 76
+LL+ + +GS GP++ V+ E V ++ T++ Y +EGR P+L ND S F L+
Sbjct: 545 KLLLKVTPMGSLGPVQVVMTPESTVGDLVAVTVRQYVKEGRRPILPSNDPSHFDLHYSQF 724
Query: 77 GSDAALSPWDKIGSHGARN 95
S +L +K+ G+RN
Sbjct: 725 -SLQSLDREEKLMDIGSRN 778
>TC207265 similar to UP|O49657 (O49657) Predicted protein, partial (6%)
Length = 796
Score = 39.7 bits (91), Expect = 5e-04
Identities = 25/89 (28%), Positives = 43/89 (48%)
Frame = +3
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGNDHSAFFLYCPHLG 77
++++++ V GS GP+R +V VE I + Y EGR P + + ++ F
Sbjct: 48 KVVVNVTVEGSPGPVRTMVKLGSSVEDTIKRVVDKYTEEGRSPQIDPNMASSFQLHHSYF 227
Query: 78 SDAALSPWDKIGSHGARNFVLCKKPQAAT 106
S +L I G+R+F L K A++
Sbjct: 228 SLQSLDKSQVIADVGSRSFYLRKNNDASS 314
>TC207266 weakly similar to UP|O49657 (O49657) Predicted protein, partial
(9%)
Length = 1036
Score = 38.5 bits (88), Expect = 0.001
Identities = 24/89 (26%), Positives = 44/89 (48%)
Frame = +2
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGNDHSAFFLYCPHLG 77
++++++ V GS GP++ +V VE I + Y EGR P + + ++ F
Sbjct: 605 KVVVNVTVEGSPGPVQTMVKLGSSVEDTIKRVVDKYTEEGRSPQIDANMASSFQLHHSYF 784
Query: 78 SDAALSPWDKIGSHGARNFVLCKKPQAAT 106
S +L + I G+R+F L K A++
Sbjct: 785 SLHSLDKSEVIADVGSRSFYLRKNNDASS 871
>TC225720
Length = 800
Score = 35.4 bits (80), Expect = 0.009
Identities = 13/45 (28%), Positives = 27/45 (59%)
Frame = +3
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVL 62
++L+ + +L S P++ ++ E V +I ++ Y +EGR P+L
Sbjct: 429 KMLLKVTMLRSLAPVQMLMTPESTVRDLIAAAVRQYVKEGRRPIL 563
>BM525705 weakly similar to GP|2827553|emb| predicted protein {Arabidopsis
thaliana}, partial (4%)
Length = 435
Score = 34.3 bits (77), Expect = 0.020
Identities = 16/53 (30%), Positives = 29/53 (54%)
Frame = +1
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGNDHSAFF 70
++++++ V GS GP+R +V VE I + Y EGR P + + ++ F
Sbjct: 157 KVVVNVTVEGSPGPVRTMVKLGSSVEDTIKRVVDKYTEEGRSPQIDPNMASSF 315
>TC225721 homologue to UP|Q7S1C8 (Q7S1C8) Predicted protein, partial (13%)
Length = 1010
Score = 33.9 bits (76), Expect = 0.026
Identities = 14/53 (26%), Positives = 29/53 (54%)
Frame = +2
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGNDHSAFF 70
++L+ + VL S P++ ++ V +I ++ Y +EGR P+L + + F
Sbjct: 89 KMLLKVTVLRSLAPVQVLMTPGSTVRDLIAAAVRQYVKEGRRPILPSSEVSEF 247
>TC212539
Length = 321
Score = 32.7 bits (73), Expect = 0.057
Identities = 18/48 (37%), Positives = 30/48 (62%), Gaps = 2/48 (4%)
Frame = +1
Query: 95 NFVLCKKPQAATNEAAAEDGSGTSSLPRRGSGSWKSWFN--LNLKVSS 140
NFVLCKK Q +++AA + + + ++ +G WK+W N +LK+ S
Sbjct: 4 NFVLCKK-QVQSSKAAPQ----SELVSQKSNGGWKAWLNKSFSLKILS 132
>BF008686
Length = 349
Score = 31.6 bits (70), Expect = 0.13
Identities = 25/103 (24%), Positives = 46/103 (44%), Gaps = 1/103 (0%)
Frame = +2
Query: 20 LISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGN-DHSAFFLYCPHLGS 78
L+ ++ +GS GP+ V++ V + ++ + R P+L + D S F L+
Sbjct: 2 LLKVSSMGSLGPVLVVLSPVSSVGVFVAAAVRLFV*VSRRPILPSFDPSLFVLHYSQFSL 181
Query: 79 DAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSLP 121
+ +L +K+ G+R F +C + G G SS P
Sbjct: 182 E-SLDREEKLRDIGSRIFFMCPM------KCGGGGGVGCSSSP 289
>TC234963 homologue to UP|Q9FH57 (Q9FH57) GATA-binding transcription
factor-like protein (At5g66320), partial (8%)
Length = 387
Score = 28.9 bits (63), Expect = 0.83
Identities = 27/95 (28%), Positives = 42/95 (43%), Gaps = 5/95 (5%)
Frame = -2
Query: 46 IDTTLKSYAREGRLPVLGNDHSAFFLYCPHLGSDAALSPWDKIGSHGARNFVLC-----K 100
+ T + +Y + RL + G +Y + G S W + S+ F L +
Sbjct: 308 LTTNINTYHNQRRLKLKGKK-----MYTLNPGPH*NRSIWSNLFSNYYSFFFLT*KKKIQ 144
Query: 101 KPQAATNEAAAEDGSGTSSLPRRGSGSWKSWFNLN 135
KP + EA + GSGTSS RR S +++ W N
Sbjct: 143 KPGSDGGEAVS--GSGTSSFFRRISRTFRWWLECN 45
>TC220147
Length = 1630
Score = 27.7 bits (60), Expect = 1.8
Identities = 12/23 (52%), Positives = 17/23 (73%)
Frame = -1
Query: 4 SKQKKNQNANNAKRRLLISINVL 26
SK +KNQNAN A ++ L+ NV+
Sbjct: 1477 SKWQKNQNANYALQKFLVQTNVV 1409
>TC229748
Length = 914
Score = 26.9 bits (58), Expect = 3.1
Identities = 11/18 (61%), Positives = 11/18 (61%)
Frame = -3
Query: 114 GSGTSSLPRRGSGSWKSW 131
GSG SS RRG G W W
Sbjct: 330 GSGKSSETRRGVGEWFEW 277
>CD411677
Length = 623
Score = 26.6 bits (57), Expect = 4.1
Identities = 12/34 (35%), Positives = 18/34 (52%), Gaps = 1/34 (2%)
Frame = +2
Query: 59 LPVLGNDHSAFFLYCPHLGSDAALSPW-DKIGSH 91
L L N H FL+ H+ ++ W +K+GSH
Sbjct: 26 LKPLQNSHRYLFLFTVHITKSPSILSWSEKLGSH 127
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,546,542
Number of Sequences: 63676
Number of extensions: 83706
Number of successful extensions: 402
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 395
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 395
length of query: 140
length of database: 12,639,632
effective HSP length: 88
effective length of query: 52
effective length of database: 7,036,144
effective search space: 365879488
effective search space used: 365879488
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 54 (25.4 bits)
Medicago: description of AC146775.17


