

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146775.16 - phase: 0
(172 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
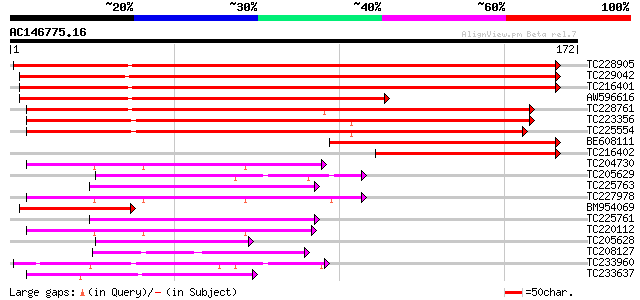
Score E
Sequences producing significant alignments: (bits) Value
TC228905 weakly similar to UP|Q7XIV3 (Q7XIV3) Zinc finger and C2... 274 2e-74
TC229042 similar to UP|Q9LVH4 (Q9LVH4) GTPase activating protein... 202 5e-53
TC216401 weakly similar to UP|Q9LP65 (Q9LP65) T1N15.21, partial ... 201 1e-52
AW596616 120 2e-28
TC228761 similar to GB|AAP68261.1|31711810|BT008822 At4g21160 {A... 119 9e-28
TC223356 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {A... 117 3e-27
TC225554 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {A... 114 2e-26
BE608111 114 2e-26
TC216402 similar to UP|Q6YWF1 (Q6YWF1) C2 domain-containing prot... 70 5e-13
TC204730 similar to UP|Q8LQF0 (Q8LQF0) OJ1529_G03.6 protein, par... 55 2e-08
TC205629 similar to UP|Q9LEX1 (Q9LEX1) CaLB protein (At3g61050),... 52 2e-07
TC225763 50 4e-07
TC227978 similar to GB|CAE85115.1|39918793|AJ617630 synaptotagmi... 49 9e-07
BM954069 49 9e-07
TC225761 weakly similar to UP|O50006 (O50006) Os-FIERG2 protein,... 49 2e-06
TC220112 similar to UP|Q7XA06 (Q7XA06) Synaptotagmin C, partial ... 47 6e-06
TC205628 similar to UP|O48645 (O48645) CLB1, partial (38%) 45 2e-05
TC208127 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8,... 45 2e-05
TC233960 similar to UP|OPSD_SEPOF (O16005) Rhodopsin, partial (10%) 41 2e-04
TC233637 40 5e-04
>TC228905 weakly similar to UP|Q7XIV3 (Q7XIV3) Zinc finger and C2 domain
protein-like, partial (85%)
Length = 819
Score = 274 bits (700), Expect = 2e-74
Identities = 129/166 (77%), Positives = 152/166 (90%)
Frame = +3
Query: 2 DNNVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLS 61
+NN+LGLLKLRIKRG+NLAIRD+ +SDPYVVVN+G +QKLKTRVVKNNCNP+WNEELTLS
Sbjct: 42 NNNILGLLKLRIKRGVNLAIRDARTSDPYVVVNMG-DQKLKTRVVKNNCNPDWNEELTLS 218
Query: 62 IRDVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTN 121
++DV+ PI LTV+DKDTF VDDKMG+AEIDLKPY QC +M L LPNGC++KR+Q +RTN
Sbjct: 219 VKDVKTPIHLTVYDKDTFSVDDKMGEAEIDLKPYVQCKQMGLGKLPNGCSLKRIQPDRTN 398
Query: 122 CLAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
LAEESSCIW+NGK++QEM LRLRNVESGE++VEIEWVDV GCKGL
Sbjct: 399 YLAEESSCIWQNGKIVQEMFLRLRNVESGEILVEIEWVDVVGCKGL 536
>TC229042 similar to UP|Q9LVH4 (Q9LVH4) GTPase activating protein-like,
partial (95%)
Length = 783
Score = 202 bits (515), Expect = 5e-53
Identities = 94/164 (57%), Positives = 129/164 (78%)
Frame = +3
Query: 4 NVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIR 63
N+LGLL++R+KRG+NLA+RD SSDPYVV+ + + QKLKTRV+K + NPEWNE+LTLS+
Sbjct: 24 NLLGLLRVRVKRGVNLAVRDVRSSDPYVVIKM-YNQKLKTRVIKKDVNPEWNEDLTLSVI 200
Query: 64 DVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTNCL 123
+ I LTV+D DTF DDKMGDAE D+ P+ + +KM L LPNG + R+Q ++ NCL
Sbjct: 201 NPNHKIKLTVYDHDTFSKDDKMGDAEFDIFPFIEALKMNLTGLPNGTVVTRIQPSKHNCL 380
Query: 124 AEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
A+ES + NGKV+Q+MILRL+NVE GE+ ++++W+D+PG KGL
Sbjct: 381 ADESCITYSNGKVVQDMILRLQNVECGEVEIQLQWIDLPGSKGL 512
>TC216401 weakly similar to UP|Q9LP65 (Q9LP65) T1N15.21, partial (82%)
Length = 944
Score = 201 bits (511), Expect = 1e-52
Identities = 94/164 (57%), Positives = 128/164 (77%)
Frame = +3
Query: 4 NVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIR 63
N+LGLL++ +++G+NLAIRD SSDPYVV+ +G +QKLKTRVV N NPEWN++LTLSI
Sbjct: 93 NLLGLLRIHVEKGVNLAIRDVVSSDPYVVIKMG-KQKLKTRVVNKNLNPEWNDDLTLSIS 269
Query: 64 DVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTNCL 123
D PI L V+DKDTF +DDKMGDAE + P+ + VKM+L +LPN + +V +R N L
Sbjct: 270 DPHAPIHLHVYDKDTFSMDDKMGDAEFFIGPFIEAVKMRLSSLPNNTIVTKVLPSRQNSL 449
Query: 124 AEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
AEES +WK+GKV+Q M+LRLRNVE+GE+ +++ W+D+PG + L
Sbjct: 450 AEESHIVWKDGKVVQNMVLRLRNVETGEVELQLHWIDIPGSRHL 581
>AW596616
Length = 337
Score = 120 bits (302), Expect = 2e-28
Identities = 59/112 (52%), Positives = 81/112 (71%)
Frame = +2
Query: 4 NVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIR 63
N+LGLL++ +++G+NLAIRD S+DPYVV+ +G + KLKTRVV N NPEWN++LTLSI
Sbjct: 5 NLLGLLRIHVEKGVNLAIRDVVSTDPYVVIKMG-KSKLKTRVVDKNLNPEWNDDLTLSIS 181
Query: 64 DVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRV 115
D PI L V+DKDTF +DDKM DA + + P+ V+++ LPN + V
Sbjct: 182 DPHAPIHLHVYDKDTFIMDDKMVDAYLFISPFM*TVEIR*SNLPNNTIVTEV 337
>TC228761 similar to GB|AAP68261.1|31711810|BT008822 At4g21160 {Arabidopsis
thaliana;} , partial (48%)
Length = 729
Score = 119 bits (297), Expect = 9e-28
Identities = 66/155 (42%), Positives = 101/155 (64%), Gaps = 1/155 (0%)
Frame = +1
Query: 6 LGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
+G+LK+++ +G NLAIRD SSDPYVV+++G +Q ++T ++++N NP WNEE LS+ +
Sbjct: 31 IGMLKVKVIKGTNLAIRDIKSSDPYVVLSLG-QQTVQTTIIRSNLNPVWNEEYMLSVPEH 207
Query: 66 RVPICLTVFDKDTFFVDDKMGDAEIDLKP-YTQCVKMKLDTLPNGCAIKRVQANRTNCLA 124
+ L VFD DTF DD MG+A+IDL+ T + + I + + N L
Sbjct: 208 YGQMKLKVFDHDTFSADDIMGEADIDLQSLITSAMAFGDAGMFGNMQIGKWLKSDDNALI 387
Query: 125 EESSCIWKNGKVLQEMILRLRNVESGELVVEIEWV 159
E+S+ +GKV Q M L+L++VESGEL +E+EW+
Sbjct: 388 EDSTVNIVDGKVKQMMSLKLQDVESGELDLELEWI 492
>TC223356 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {Arabidopsis
thaliana;} , partial (96%)
Length = 705
Score = 117 bits (293), Expect = 3e-27
Identities = 67/156 (42%), Positives = 97/156 (61%), Gaps = 2/156 (1%)
Frame = +3
Query: 6 LGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
L +LK+ + +G L IRD SSDPYVVV +G+ Q KTRV++ NP WNEEL ++ +
Sbjct: 78 LKILKVIVVQGKRLVIRDFKSSDPYVVVKLGN-QTAKTRVIRCCLNPVWNEELNFTLTEP 254
Query: 66 RVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMK--LDTLPNGCAIKRVQANRTNCL 123
+ L VFDKD + DDKMG++ ++L+P +++ L +++V + NCL
Sbjct: 255 LGVLNLEVFDKDLWKADDKMGNSYLNLQPLISAARLRDILKVSSGETTLRKVTPDSENCL 434
Query: 124 AEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWV 159
A ESS NG+VLQ + LRLR VESGEL + I+ +
Sbjct: 435 ARESSINCVNGEVLQNVWLRLRGVESGELQLTIKLI 542
>TC225554 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {Arabidopsis
thaliana;} , partial (95%)
Length = 1383
Score = 114 bits (286), Expect = 2e-26
Identities = 65/154 (42%), Positives = 95/154 (61%), Gaps = 2/154 (1%)
Frame = +3
Query: 6 LGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
LGLLK+ + +G L IRD +SDPYVV+ +G+ Q KT+V+ + NP WNEEL ++ +
Sbjct: 297 LGLLKVIVVQGKRLVIRDFKTSDPYVVLKLGN-QTAKTKVINSCLNPVWNEELNFTLTEP 473
Query: 66 RVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMK--LDTLPNGCAIKRVQANRTNCL 123
+ L VFDKD DDKMG+A ++L+P +++ L +++V + NCL
Sbjct: 474 LGVLNLEVFDKDLLKADDKMGNAFLNLQPIVSAARLRDILRVSSGETTLRKVIPDGENCL 653
Query: 124 AEESSCIWKNGKVLQEMILRLRNVESGELVVEIE 157
ESS NG+V+Q + LRLR VESGEL + I+
Sbjct: 654 VRESSINCVNGEVVQNVWLRLRGVESGELELTIK 755
>BE608111
Length = 422
Score = 114 bits (286), Expect = 2e-26
Identities = 51/70 (72%), Positives = 62/70 (87%)
Frame = -3
Query: 98 CVKMKLDTLPNGCAIKRVQANRTNCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEIE 157
C +M L LPNGC++KR+Q +RTN LAEESSCIW+NGK++QEM LRLRNVESGE++VEIE
Sbjct: 420 CKQMGLGKLPNGCSLKRIQPDRTNYLAEESSCIWQNGKIVQEMFLRLRNVESGEILVEIE 241
Query: 158 WVDVPGCKGL 167
WVDV GC+GL
Sbjct: 240 WVDVVGCRGL 211
>TC216402 similar to UP|Q6YWF1 (Q6YWF1) C2 domain-containing protein-like,
partial (35%)
Length = 497
Score = 70.1 bits (170), Expect = 5e-13
Identities = 30/56 (53%), Positives = 44/56 (78%)
Frame = +2
Query: 112 IKRVQANRTNCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGL 167
+ +V +R NCLAEES + K+GKV+Q M+LRLRNVESGE+ +++ W+D+PG + L
Sbjct: 20 VTKVLPSRQNCLAEESHIMLKDGKVVQNMVLRLRNVESGEVELQLHWIDIPGSRHL 187
>TC204730 similar to UP|Q8LQF0 (Q8LQF0) OJ1529_G03.6 protein, partial (97%)
Length = 2084
Score = 54.7 bits (130), Expect = 2e-08
Identities = 35/95 (36%), Positives = 51/95 (52%), Gaps = 4/95 (4%)
Frame = +1
Query: 6 LGLLKLRIKRGINLAIRDS-NSSDPYVVVNIGHEQ--KLKTRVVKNNCNPEWNEELTLSI 62
+G+L +++ R L +D +SDPYV + + E+ KT V N NPEWNEE + +
Sbjct: 976 VGILHVKVVRAEKLKKKDLLGASDPYVKLKLTEEKLPSKKTTVKYKNLNPEWNEEFNIVV 1155
Query: 63 RDVRVPIC-LTVFDKDTFFVDDKMGDAEIDLKPYT 96
+D + LTV+D + DKMG I LK T
Sbjct: 1156 KDPESQVLELTVYDWEQIGKHDKMGMNVIPLKEIT 1260
>TC205629 similar to UP|Q9LEX1 (Q9LEX1) CaLB protein (At3g61050), partial
(92%)
Length = 1766
Score = 51.6 bits (122), Expect = 2e-07
Identities = 36/86 (41%), Positives = 50/86 (57%), Gaps = 4/86 (4%)
Frame = +1
Query: 27 SDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRV-PICLTVFDKDTFFVDDKM 85
SDPYVVV+I K KT+V+ NN NP WNE+ L D + L V DKD D ++
Sbjct: 760 SDPYVVVHIRPLFKYKTKVIDNNLNPTWNEKFELIAEDKETQSLILEVLDKD-IGQDKRL 936
Query: 86 GDAE---IDLKPYTQCVKMKLDTLPN 108
G A+ IDL+ T+ +++L LP+
Sbjct: 937 GIAQLPLIDLEIQTE-KEIELRLLPS 1011
>TC225763
Length = 847
Score = 50.4 bits (119), Expect = 4e-07
Identities = 24/70 (34%), Positives = 36/70 (51%)
Frame = +2
Query: 25 NSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICLTVFDKDTFFVDDK 84
+S DPYV++ ++K T P+WNE ++ D + L + DKD F DD
Sbjct: 215 SSIDPYVILTYRAQEKKSTVQEDAGSKPQWNESFLFTVSDSASELNLKIMDKDNFSQDDC 394
Query: 85 MGDAEIDLKP 94
+G+A I L P
Sbjct: 395 LGEATIHLDP 424
>TC227978 similar to GB|CAE85115.1|39918793|AJ617630 synaptotagmin
{Arabidopsis thaliana;} , partial (72%)
Length = 1470
Score = 49.3 bits (116), Expect = 9e-07
Identities = 33/109 (30%), Positives = 57/109 (52%), Gaps = 6/109 (5%)
Frame = +2
Query: 6 LGLLKLRIKRGINLAIRDS-NSSDPYVVVNIGHEQ--KLKTRVVKNNCNPEWNEELTLSI 62
+G+L +++ + + L +D +SDPYV + + ++ KT V N NP+WNEE + +
Sbjct: 374 VGILHVKVLQAMKLKKKDLLGASDPYVKLKLTEDKWPSKKTTVKHKNLNPQWNEEFNIVV 553
Query: 63 RDVRVPIC-LTVFDKDTFFVDDKMGDAEIDLKPYT--QCVKMKLDTLPN 108
+D + + V+D + DKMG I LK + + + LD L N
Sbjct: 554 KDPDSQVIEINVYDWEQVGKHDKMGMNVIPLKEVSPEETKRFTLDLLKN 700
>BM954069
Length = 421
Score = 49.3 bits (116), Expect = 9e-07
Identities = 20/35 (57%), Positives = 30/35 (85%)
Frame = +3
Query: 4 NVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHE 38
N+LGLL++ +++G+NLAIRD SSDPYVV+ +G +
Sbjct: 69 NLLGLLRIHVEKGVNLAIRDVVSSDPYVVIKMGRQ 173
>TC225761 weakly similar to UP|O50006 (O50006) Os-FIERG2 protein, partial
(35%)
Length = 819
Score = 48.5 bits (114), Expect = 2e-06
Identities = 24/70 (34%), Positives = 35/70 (49%)
Frame = +1
Query: 25 NSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICLTVFDKDTFFVDDK 84
+S DPYV++ ++K T P+WNE ++ D + L + DKD F DD
Sbjct: 220 SSIDPYVILTYRAQEKKSTVQEDAGSKPQWNESFLFTVSDSASELNLKIMDKDNFSQDDC 399
Query: 85 MGDAEIDLKP 94
+G A I L P
Sbjct: 400 LGVATIHLDP 429
>TC220112 similar to UP|Q7XA06 (Q7XA06) Synaptotagmin C, partial (76%)
Length = 1867
Score = 46.6 bits (109), Expect = 6e-06
Identities = 30/92 (32%), Positives = 50/92 (53%), Gaps = 4/92 (4%)
Frame = +2
Query: 6 LGLLKLRIKRGINLAIRDS-NSSDPYVVVNIGHEQ--KLKTRVVKNNCNPEWNEELTLSI 62
+G+L + + R L D +SDPYV +++ ++ KT V + N NPEWNE+ + +
Sbjct: 875 VGILHVNVVRAQKLLKMDLLGTSDPYVKLSLTGDKLPAKKTTVKRKNLNPEWNEKFKIVV 1054
Query: 63 RDVRVPIC-LTVFDKDTFFVDDKMGDAEIDLK 93
+D + + L V+D D DK+G + LK
Sbjct: 1055KDPQSQVLQLQVYDWDKVGGHDKLGMQLVPLK 1150
Score = 33.1 bits (74), Expect = 0.066
Identities = 23/92 (25%), Positives = 45/92 (48%), Gaps = 5/92 (5%)
Frame = +2
Query: 7 GLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRD-- 64
GLL + I+ + + ++P+ V+ E+K +T+ +K +P WNEE + +
Sbjct: 1373 GLLSIVIQEAEE--VEGDHHNNPFAVLTFRGEKK-RTKTMKKTRHPRWNEEFQFMLEEPP 1543
Query: 65 VRVPICLTVFDKD---TFFVDDKMGDAEIDLK 93
+ I + V K +F + +G EI+L+
Sbjct: 1544 LHEKIHIEVMSKRKNFSFLPKESLGHVEINLR 1639
>TC205628 similar to UP|O48645 (O48645) CLB1, partial (38%)
Length = 814
Score = 45.1 bits (105), Expect = 2e-05
Identities = 22/48 (45%), Positives = 28/48 (57%)
Frame = +2
Query: 27 SDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICLTVF 74
SDPYVVV+I K KT+V+ NN NP WNE+ L D + + F
Sbjct: 161 SDPYVVVHIRPLFKYKTKVIDNNLNPIWNEKFKLIAEDKETQLLILRF 304
>TC208127 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8, partial
(23%)
Length = 845
Score = 44.7 bits (104), Expect = 2e-05
Identities = 23/66 (34%), Positives = 37/66 (55%)
Frame = +3
Query: 26 SSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICLTVFDKDTFFVDDKM 85
+SDPYV VN G+ +K +T+V+ NP WN+ TL D P+ L V D + + +
Sbjct: 312 TSDPYVRVNYGNSKK-RTKVIHKTLNPRWNQ--TLEFLDDGSPLILHVKDHNALLPESSI 482
Query: 86 GDAEID 91
G+ ++
Sbjct: 483 GEGVVE 500
>TC233960 similar to UP|OPSD_SEPOF (O16005) Rhodopsin, partial (10%)
Length = 862
Score = 41.2 bits (95), Expect = 2e-04
Identities = 35/102 (34%), Positives = 51/102 (49%), Gaps = 6/102 (5%)
Frame = +2
Query: 2 DNNVLGLLKLRIKRGINLAIRD-SNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTL 60
DN+ L L K+R+ +L D +SDP+V GH +KT V NP WNE+L
Sbjct: 71 DNSPLKL-KIRVISASSLQAADRGGTSDPFVHFKQGH-MGVKTTVKNKELNPVWNEDLQF 244
Query: 61 SI--RDVRV-PICLTVFDKDTFFVDDKMGDAEIDLK--PYTQ 97
I +D + + + V DKD DD +G ++ L P+ Q
Sbjct: 245 GIYEKDAQTGTLSIEVLDKD-MLSDDCIGTYKVSLSEVPHNQ 367
>TC233637
Length = 426
Score = 40.0 bits (92), Expect = 5e-04
Identities = 23/73 (31%), Positives = 39/73 (52%), Gaps = 3/73 (4%)
Frame = +1
Query: 6 LGLLKLRIKRGINLA---IRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSI 62
+G+L+L I NL R+ ++D Y V G++ ++TR + + +P WNE+ T +
Sbjct: 178 IGILELGILSARNLLPMKAREGRTTDAYCVAKYGNKW-VRTRTLLDTLSPRWNEQYTWEV 354
Query: 63 RDVRVPICLTVFD 75
D I + VFD
Sbjct: 355 HDPCTVITVGVFD 393
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,814,712
Number of Sequences: 63676
Number of extensions: 100810
Number of successful extensions: 456
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 444
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 445
length of query: 172
length of database: 12,639,632
effective HSP length: 91
effective length of query: 81
effective length of database: 6,845,116
effective search space: 554454396
effective search space used: 554454396
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146775.16


