

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146758.2 + phase: 0
(74 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
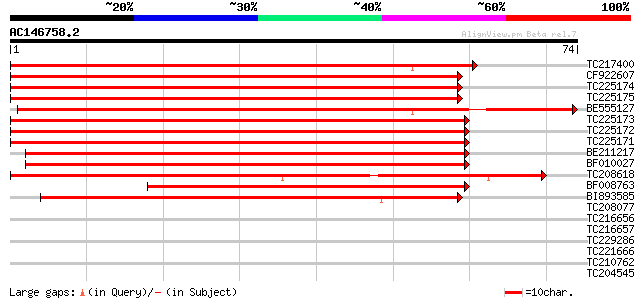
Score E
Sequences producing significant alignments: (bits) Value
TC217400 weakly similar to UP|Q9SWS4 (Q9SWS4) Ripening related p... 93 3e-20
CF922607 92 5e-20
TC225174 similar to UP|Q9SWS4 (Q9SWS4) Ripening related protein,... 92 5e-20
TC225175 similar to UP|Q9SWS4 (Q9SWS4) Ripening related protein,... 91 1e-19
BE555127 90 2e-19
TC225173 similar to UP|Q9SWS4 (Q9SWS4) Ripening related protein,... 89 4e-19
TC225172 similar to UP|Q9SWS4 (Q9SWS4) Ripening related protein,... 89 5e-19
TC225171 UP|Q9SWS4 (Q9SWS4) Ripening related protein, complete 85 8e-18
BE211217 weakly similar to GP|5739198|gb| ripening related prote... 69 6e-13
BF010027 similar to GP|5739198|gb|A ripening related protein {Gl... 65 8e-12
TC208618 weakly similar to UP|Q9SWS4 (Q9SWS4) Ripening related p... 60 2e-10
BF008763 similar to GP|5739198|gb| ripening related protein {Gly... 55 5e-09
BI893585 48 1e-06
TC208077 weakly similar to UP|Q9SXL8 (Q9SXL8) Csf-2 protein, par... 36 0.003
TC216656 weakly similar to UP|MHK_ARATH (P43294) Serine/threonin... 32 0.060
TC216657 similar to UP|MHK_ARATH (P43294) Serine/threonine-prote... 31 0.13
TC229286 weakly similar to UP|Q9SXL8 (Q9SXL8) Csf-2 protein, par... 28 0.87
TC221666 27 1.5
TC210762 27 2.5
TC204545 homologue to UP|Q9LKY8 (Q9LKY8) Proline-rich protein, c... 27 2.5
>TC217400 weakly similar to UP|Q9SWS4 (Q9SWS4) Ripening related protein,
partial (98%)
Length = 800
Score = 92.8 bits (229), Expect = 3e-20
Identities = 41/62 (66%), Positives = 51/62 (82%), Gaps = 1/62 (1%)
Frame = +1
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHH-SDSVKHWT 59
MVLAGKLSTE+G+KAS+ K+F L + +H VQN+CE VHETKLH GD+WH +SVKHWT
Sbjct: 64 MVLAGKLSTELGVKASATKWFNLLTTQLHHVQNLCERVHETKLHDGDDWHGVGNSVKHWT 243
Query: 60 HV 61
+V
Sbjct: 244 YV 249
>CF922607
Length = 560
Score = 92.0 bits (227), Expect = 5e-20
Identities = 38/59 (64%), Positives = 49/59 (82%)
Frame = -3
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWT 59
M LAGK+S EIG+ AS+ K+F LFA+ +H VQN+ E VHETKLH GD+WHH++S+KHWT
Sbjct: 486 MSLAGKISIEIGVHASAAKWFNLFATQLHHVQNLTERVHETKLHHGDDWHHNESIKHWT 310
>TC225174 similar to UP|Q9SWS4 (Q9SWS4) Ripening related protein, complete
Length = 834
Score = 92.0 bits (227), Expect = 5e-20
Identities = 38/59 (64%), Positives = 49/59 (82%)
Frame = +3
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWT 59
M LAGK+S EIG+ AS+ K+F LFA+ +H VQN+ E VHETKLH GD+WHH++S+KHWT
Sbjct: 72 MSLAGKISIEIGVHASAAKWFNLFATQLHHVQNLTERVHETKLHHGDDWHHNESIKHWT 248
>TC225175 similar to UP|Q9SWS4 (Q9SWS4) Ripening related protein, complete
Length = 784
Score = 90.5 bits (223), Expect = 1e-19
Identities = 36/59 (61%), Positives = 49/59 (83%)
Frame = +2
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWT 59
M L GK+STEIG+ A++ K+F LFA+ +H VQN+ + VHETKLH GD+WHH++S+KHWT
Sbjct: 56 MSLVGKISTEIGVHATAAKWFNLFATQLHHVQNLTDRVHETKLHHGDDWHHNESIKHWT 232
>BE555127
Length = 442
Score = 89.7 bits (221), Expect = 2e-19
Identities = 42/74 (56%), Positives = 54/74 (72%), Gaps = 1/74 (1%)
Frame = +2
Query: 2 VLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHH-SDSVKHWTH 60
VLAGKLSTE+G+KA + K+F L + +H VQN+CE VHETKLH GD+WH +SVKHWT+
Sbjct: 2 VLAGKLSTELGVKAFATKWFNLLTTQLHHVQNLCERVHETKLHDGDDWHGVGNSVKHWTY 181
Query: 61 VRVLSSKQRLLHKS 74
V+ K H+S
Sbjct: 182--VIDGKVHTCHES 217
>TC225173 similar to UP|Q9SWS4 (Q9SWS4) Ripening related protein, partial
(96%)
Length = 784
Score = 89.0 bits (219), Expect = 4e-19
Identities = 35/60 (58%), Positives = 51/60 (84%)
Frame = +2
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTH 60
M LAGK++TEIG+ A++ K+F LFA+ +H VQN+ + VH TKLHQG++WHH+++VKHWT+
Sbjct: 65 MSLAGKITTEIGVHATATKWFNLFATQLHHVQNLTDRVHGTKLHQGEDWHHNETVKHWTY 244
>TC225172 similar to UP|Q9SWS4 (Q9SWS4) Ripening related protein, complete
Length = 971
Score = 88.6 bits (218), Expect = 5e-19
Identities = 35/60 (58%), Positives = 51/60 (84%)
Frame = +1
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTH 60
M LAGK++TEIG+ A++ K+F LFA+ +H VQN+ + VH TKLHQG++WHH+++VKHWT+
Sbjct: 271 MSLAGKITTEIGVHATAAKWFNLFATQLHHVQNLTDRVHGTKLHQGEDWHHNETVKHWTY 450
>TC225171 UP|Q9SWS4 (Q9SWS4) Ripening related protein, complete
Length = 782
Score = 84.7 bits (208), Expect = 8e-18
Identities = 32/60 (53%), Positives = 50/60 (83%)
Frame = +1
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTH 60
M LAGK+S+EIG++A++ K+F LF + +H VQN+ + +H T+LH G++WHH++SVKHWT+
Sbjct: 76 MSLAGKISSEIGVQATAAKWFNLFTTQLHHVQNLTDRIHGTELHHGEDWHHNESVKHWTY 255
>BE211217 weakly similar to GP|5739198|gb| ripening related protein
{Glycine max}, partial (98%)
Length = 454
Score = 68.6 bits (166), Expect = 6e-13
Identities = 30/58 (51%), Positives = 44/58 (75%)
Frame = +2
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTH 60
LAGK++TEIG+ A++ K+F LFA+ + VQN+ + VH TKLH G+ H+ +VKHWT+
Sbjct: 5 LAGKITTEIGVHATAAKWFNLFATQLLHVQNLSDRVHGTKLH*GENCLHNKTVKHWTY 178
>BF010027 similar to GP|5739198|gb|A ripening related protein {Glycine
max}, partial (54%)
Length = 256
Score = 64.7 bits (156), Expect = 8e-12
Identities = 26/58 (44%), Positives = 44/58 (75%)
Frame = +2
Query: 3 LAGKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTH 60
LAG++ +EIG++A++ K F LF +++H VQN+ + +H T+L + HH++SVKHWT+
Sbjct: 5 LAGEIISEIGVQATAAKRFNLFTTHLHHVQNLTDKIHGTELDLDEY*HHNESVKHWTY 178
>TC208618 weakly similar to UP|Q9SWS4 (Q9SWS4) Ripening related protein,
partial (26%)
Length = 808
Score = 60.5 bits (145), Expect = 2e-10
Identities = 32/74 (43%), Positives = 47/74 (63%), Gaps = 4/74 (5%)
Frame = +3
Query: 1 MVLAGKLSTEIGIKASSDKFFKLFASNIHEVQNI-CESVHETKLHQGDEWHHSDSVKHWT 59
MVL+GK+ TE+ I+A + KF+ +F IH V N+ E VH K+H+GD W + SVKHW
Sbjct: 54 MVLSGKVETEVEIQAPAAKFYHVFRKQIHHVPNMSTERVHGAKVHEGD-WENIGSVKHWD 230
Query: 60 HV---RVLSSKQRL 70
R S+K+++
Sbjct: 231 FTIEGRKTSAKEKI 272
>BF008763 similar to GP|5739198|gb| ripening related protein {Glycine max},
partial (70%)
Length = 409
Score = 55.5 bits (132), Expect = 5e-09
Identities = 20/42 (47%), Positives = 31/42 (73%)
Frame = +2
Query: 19 KFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTH 60
++F LFA+ +H VQN+ + +H TKLH G++ HH +V HWT+
Sbjct: 11 QWFNLFATQLHHVQNLTDKLHGTKLHHGEDCHHIHTVNHWTY 136
>BI893585
Length = 421
Score = 47.8 bits (112), Expect = 1e-06
Identities = 22/56 (39%), Positives = 34/56 (60%), Gaps = 1/56 (1%)
Frame = -2
Query: 5 GKLSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGD-EWHHSDSVKHWT 59
GK TE+ I++ + KFF +A + QNI ++ H++KL + +WH SVK WT
Sbjct: 408 GKAVTELEIQSPTVKFFNAYAKELQNGQNIVDTRHDSKLQDNERDWHDFGSVKSWT 241
>TC208077 weakly similar to UP|Q9SXL8 (Q9SXL8) Csf-2 protein, partial (72%)
Length = 616
Score = 36.2 bits (82), Expect = 0.003
Identities = 18/57 (31%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Frame = +1
Query: 6 KLSTEIGIKASSDKFFKLFASNIHEVQNICE-SVHETKLHQGDEWHHSDSVKHWTHV 61
K+ + IKAS+D+F + + H + NI V ++H+G EW SV W ++
Sbjct: 49 KVEANVHIKASADQFHNVLCNRTHHIANIFPGKVQSVEIHKG-EWGTEGSVISWNYL 216
>TC216656 weakly similar to UP|MHK_ARATH (P43294) Serine/threonine-protein
kinase MHK , partial (15%)
Length = 802
Score = 32.0 bits (71), Expect = 0.060
Identities = 23/68 (33%), Positives = 30/68 (43%)
Frame = +1
Query: 7 LSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTHVRVLSS 66
L +G+K S SN+ VQN+ + V E L D HSD WT +LS
Sbjct: 220 LGLTLGVKPS--------VSNLDVVQNVSQGVRENVLFCSDFNDHSDQSVFWT---LLSP 366
Query: 67 KQRLLHKS 74
Q +H S
Sbjct: 367 DQNGVHNS 390
>TC216657 similar to UP|MHK_ARATH (P43294) Serine/threonine-protein kinase
MHK , partial (41%)
Length = 1393
Score = 30.8 bits (68), Expect = 0.13
Identities = 23/68 (33%), Positives = 29/68 (41%)
Frame = +1
Query: 7 LSTEIGIKASSDKFFKLFASNIHEVQNICESVHETKLHQGDEWHHSDSVKHWTHVRVLSS 66
L +G+K S SN VQN+ + V E L D HSD WT +LS
Sbjct: 781 LGLTLGVKPS--------VSNSDVVQNVSQGVRENVLFCSDFNDHSDQSVFWT---LLSP 927
Query: 67 KQRLLHKS 74
Q +H S
Sbjct: 928 DQNGIHNS 951
>TC229286 weakly similar to UP|Q9SXL8 (Q9SXL8) Csf-2 protein, partial (34%)
Length = 759
Score = 28.1 bits (61), Expect = 0.87
Identities = 14/57 (24%), Positives = 28/57 (48%), Gaps = 1/57 (1%)
Frame = +2
Query: 6 KLSTEIGIKASSDKFFKLFASNIHEVQNIC-ESVHETKLHQGDEWHHSDSVKHWTHV 61
KL + IKAS+++F+ + H++ I E ++ +G W S+ W ++
Sbjct: 47 KLEANVSIKASAEQFYDVLCHKTHQIPKIFPEKALSVEILKG-AWGTEGSIISWNYL 214
>TC221666
Length = 485
Score = 27.3 bits (59), Expect = 1.5
Identities = 10/32 (31%), Positives = 17/32 (52%)
Frame = +2
Query: 22 KLFASNIHEVQNICESVHETKLHQGDEWHHSD 53
K F S + + E+ + K HQ ++WHH +
Sbjct: 218 KEFKSRVWNKEKTYENYIDLKWHQNNQWHHQN 313
>TC210762
Length = 857
Score = 26.6 bits (57), Expect = 2.5
Identities = 8/22 (36%), Positives = 12/22 (54%)
Frame = -3
Query: 46 GDEWHHSDSVKHWTHVRVLSSK 67
G +WHH W H ++LS +
Sbjct: 348 GAKWHHQSRASTWIHHKLLSQR 283
>TC204545 homologue to UP|Q9LKY8 (Q9LKY8) Proline-rich protein, complete
Length = 1924
Score = 26.6 bits (57), Expect = 2.5
Identities = 9/18 (50%), Positives = 11/18 (61%)
Frame = +2
Query: 43 LHQGDEWHHSDSVKHWTH 60
+H D W HS V+HW H
Sbjct: 59 VHDVDGW*HSPLVQHWDH 112
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.129 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,610,434
Number of Sequences: 63676
Number of extensions: 45061
Number of successful extensions: 351
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 346
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 346
length of query: 74
length of database: 12,639,632
effective HSP length: 50
effective length of query: 24
effective length of database: 9,455,832
effective search space: 226939968
effective search space used: 226939968
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146758.2


