

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146758.11 + phase: 0 /pseudo
(1309 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
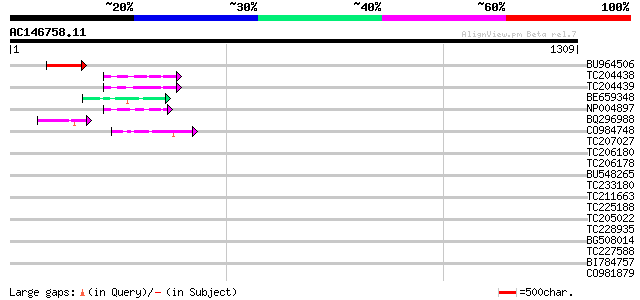
Score E
Sequences producing significant alignments: (bits) Value
BU964506 similar to PIR|C86469|C864 protein F12K21.14 [imported]... 105 1e-22
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 64 4e-10
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 62 2e-09
BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor p... 53 8e-07
NP004897 gag-protease polyprotein 50 5e-06
BQ296988 similar to GP|21740616|em OSJNBb0089K24.12 {Oryza sativ... 47 4e-05
CO984748 46 9e-05
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 42 0.001
TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 40 0.007
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 40 0.007
BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delal... 39 0.011
TC233180 similar to UP|Q944K0 (Q944K0) At1g18030/T10F20_3, parti... 39 0.015
TC211663 38 0.025
TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like prot... 37 0.074
TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, pa... 36 0.097
TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana geno... 35 0.28
BG508014 GP|15128578|db prolactin receptor {Paralichthys olivace... 34 0.48
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 34 0.48
BI784757 33 0.63
CO981879 32 1.8
>BU964506 similar to PIR|C86469|C864 protein F12K21.14 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 431
Score = 105 bits (262), Expect = 1e-22
Identities = 47/91 (51%), Positives = 71/91 (77%)
Frame = +3
Query: 86 AQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEHSAIIHADVP 145
A MWNYLK++Y Q+N A RFQLE +IA ++Q + S+ ++YS F+NLW E++ I++AD+P
Sbjct: 6 AATMWNYLKKVYGQNNEA*RFQLEHDIATFRQDSLSISDFYSQFMNLWAEYTNIVYADLP 185
Query: 146 NIYLAAVQEVYNTSKQDQFLMILRPEFEVVR 176
+ L++VQ V+ T+K+DQFLM LR +FE +R
Sbjct: 186 SEGLSSVQTVHETTKRDQFLMKLRSDFEGIR 278
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 63.9 bits (154), Expect = 4e-10
Identities = 51/185 (27%), Positives = 84/185 (44%), Gaps = 5/185 (2%)
Frame = +1
Query: 216 ESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSCSKKF-CNYCKQRGHIITECYVRPP 274
+++T A +Q R + H QQ +S KK+ C+YC + GHI CY
Sbjct: 1426 KNSTGATMSQHRSRHHGTQQ------------KKSKRKKWRCHYCGKYGHIKPFCYHLHG 1569
Query: 275 PSTQSPM*ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDSGAS 334
H + T +++ G + + ++S + ++ ++A W+LDSG S
Sbjct: 1570 ----------HPHHGTQSSSSGRKMMWVPKHKIVSLVVHTSLR---ASAKEDWYLDSGCS 1710
Query: 335 YHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDINSD----FRDVLVSPGLASNLL 390
HMTG E+L N+ + DG+ IT +G + D VL+ GL +NL+
Sbjct: 1711 RHMTGVKEFLVNIEPC-STSYVTFGDGSKGKITGMGKLVHDGLPSLNKVLLVKGLTANLI 1887
Query: 391 SVGQL 395
S+ QL
Sbjct: 1888 SISQL 1902
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 62.0 bits (149), Expect = 2e-09
Identities = 53/185 (28%), Positives = 81/185 (43%), Gaps = 5/185 (2%)
Frame = +1
Query: 216 ESTTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSCSKKF-CNYCKQRGHIITECYVRPP 274
++ T A +Q R + H MQQ +S KK+ C+YC + GHI CY
Sbjct: 1423 KNRTGATMSQHRSRHHGMQQ------------KKSKRKKWRCHYCGKYGHIKPFCY---- 1554
Query: 275 PSTQSPM*ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDSGAS 334
LH + + T S+ +M+ A+ + ++A W+LDSG S
Sbjct: 1555 --------HLHGHPH-HGTQSSNSRKKMMWVPKHKAVSLVVHTSLRASAKEDWYLDSGCS 1707
Query: 335 YHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDINSD----FRDVLVSPGLASNLL 390
HMTG E+L N+ + DG+ I +G + D VL+ GL +NL+
Sbjct: 1708 RHMTGVKEFLLNIEPC-STSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKGLTANLI 1884
Query: 391 SVGQL 395
S+ QL
Sbjct: 1885 SISQL 1899
>BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor protein
DSK-R1 - fruit fly (Drosophila melanogaster), partial
(6%)
Length = 770
Score = 53.1 bits (126), Expect = 8e-07
Identities = 56/216 (25%), Positives = 85/216 (38%), Gaps = 13/216 (6%)
Frame = -1
Query: 168 LRPEFEVVRGSLLNRNVVPSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRR 227
+ P+F+ +R +L VPSL+ ++ LLR + ++ I S V+ +
Sbjct: 602 MHPDFDHIRDQVLTSQEVPSLENLITRLLRVPSPKIGGNSVDN---IETSVMVSNRGGQG 432
Query: 228 GKGHDMQQVQCFSCKQFGHIARSCSKKFCNYCKQRGHIITECY---------VRPPPSTQ 278
G+G+ Q G R + C+YCK+ GH CY V S
Sbjct: 431 GRGN-----------QGGRAGRG-GRP*CSYCKRVGHTQDTCYSIHGFPGKSVNISKSET 288
Query: 279 SPM*ALHA----YSTTNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDSGAS 334
S + L A Y AT + S VIS S + N PW +DSGAS
Sbjct: 287 SEIKFLEADYQEYLQLKATKESQTSS------VISGHNSTACISQVGNNQSPWIIDSGAS 126
Query: 335 YHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVG 370
H+ + L I +ADG+ ++ T +G
Sbjct: 125 DHIASNSSLF*FLSPPKIPHFITLADGSRVATTGIG 18
>NP004897 gag-protease polyprotein
Length = 1923
Score = 50.4 bits (119), Expect = 5e-06
Identities = 46/162 (28%), Positives = 71/162 (43%), Gaps = 4/162 (2%)
Frame = +1
Query: 218 TTVAYAAQRRGKGHDMQQVQCFSCKQFGHIARSCSKKF-CNYCKQRGHIITECYV---RP 273
+T A +Q R + H QQ +S KK+ C+YC + GHI CY P
Sbjct: 1432 STGATMSQHRSRHHGTQQ------------KKSKRKKWRCHYCGKYGHIKPFCYHLHGHP 1575
Query: 274 PPSTQSPM*ALHAYSTTNATNGGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDSGA 333
TQS S++ V + +++ +V ++L ++A W+LDSG
Sbjct: 1576 HHGTQS--------SSSRRKMMWVPKHKIVSLVVHTSL--------RASAKEDWYLDSGC 1707
Query: 334 SYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDINSD 375
S HMTG E+L N+ + DG+ IT +G + D
Sbjct: 1708 SRHMTGVKEFLVNIEPC-STSYVTFGDGSKGKITGMGKLVHD 1830
>BQ296988 similar to GP|21740616|em OSJNBb0089K24.12 {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 408
Score = 47.4 bits (111), Expect = 4e-05
Identities = 34/133 (25%), Positives = 61/133 (45%), Gaps = 9/133 (6%)
Frame = -1
Query: 64 IITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQ 123
I +WIL+++ P + ++ A ++W LK ++Q + + +++ EI QG SV
Sbjct: 393 IHSWILNSVEPSISRSIVFMDNASDVWLDLKERFSQGDLVRVSEIQQEIYALTQGTRSVT 214
Query: 124 EYYSGFLNLWTEHSAIIHADVPNI---------YLAAVQEVYNTSKQDQFLMILRPEFEV 174
+YS LW E I+ +PN + + ++T +FL L EF
Sbjct: 213 TFYSDKKALWEELE--IYMPIPNCTCHHRCSCDAMRLARRHHHTLHVMRFLTGLNDEFNA 40
Query: 175 VRGSLLNRNVVPS 187
V+ +L +PS
Sbjct: 39 VKSQILLIEPLPS 1
>CO984748
Length = 810
Score = 46.2 bits (108), Expect = 9e-05
Identities = 54/210 (25%), Positives = 90/210 (42%), Gaps = 12/210 (5%)
Frame = -2
Query: 235 QVQCFSCKQFGHIARSCSKKFC-NYCKQRGHIITECYVRPPPSTQSPM*ALHAYSTTNAT 293
++QC +F H A C +F NY Q+ Y + Q ++Y +N
Sbjct: 806 KIQCQXXYKFDHDAADCYHRFNPNYVAQQ-------YNQNQSLNQHRN---YSYQRSNCQ 657
Query: 294 N-GGVSQSEMIQQMVISALPSIGIQGKSSNASHPWFLDSGASYHMTGSLEYLHNLHSYDG 352
SQS+ Q + L + +S N W+++ GA++H+T S + L N G
Sbjct: 656 ALNNKSQSQQNAQAPQAYLANANPTQQSQN----WYINYGATHHVTASPQNLMNEVPTSG 489
Query: 353 NKKIQIADGNTLSITDVGDINSDF--------RDVLVSPGLASNLLSVGQLVV-ILMLIF 403
N+++ + +G L IT NS F ++L P + NL+SV + F
Sbjct: 488 NEQVFLGNGQGLPITGSTVFNSPFASNVKLTLNNLLHVPHITKNLVSVS*FAKDNVFFEF 309
Query: 404 HVLVVLCKSRCRGKLSRRG-LKWEDCFLFS 432
H + KS+ G + RG L + ++FS
Sbjct: 308 HSSHCVVKSQVDGSILLRGALSQDGLYVFS 219
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 42.4 bits (98), Expect = 0.001
Identities = 41/120 (34%), Positives = 65/120 (54%), Gaps = 8/120 (6%)
Frame = -2
Query: 652 LIIFLLWFLTLTLLSFVFSKLILIIV----ICILLGVCVLFIYLRLKGISLEHNLFSVHL 707
+++FLL L L L+SF+ I+ + +C + + +LF++L L I L LF++
Sbjct: 521 ILLFLLLILFLFLISFIILTFIIFSLFL*CVCFFIFI-LLFLFLLLLFIFLFLLLFTI-- 351
Query: 708 WGIVTFIRDLCFMMSLIIASEFRGMLHFLIIN--SCFILFLLT*MIL--LFFPIFLLCLN 763
V+F L F++ I F L FL IN F+LFLL +IL +FF + LLC++
Sbjct: 350 --TVSFFFVLLFLIPFIFPIIF-FPLDFLCINFVILFLLFLLLFIILPIIFFLLDLLCIS 180
Score = 30.4 bits (67), Expect = 5.3
Identities = 30/120 (25%), Positives = 50/120 (41%), Gaps = 16/120 (13%)
Frame = -2
Query: 659 FLTLTLLSFVFSKLILIIVICILLGVCVLF------IYLRLKGISLEHNLFSVHLWGIVT 712
F L LL FVF L+L + +L F Y+ + + + + L I+
Sbjct: 713 FFLLLLLVFVFFFLLLFHFLPTILSFIFFFTFFFYFFYINIPNRNFNLAILFLTLLFILL 534
Query: 713 FIRDLCFMMSLIIASEFRGMLHFLIINS----------CFILFLLT*MILLFFPIFLLCL 762
F+ + + LI+ F ++ F+I+ CF +F+L + LL IFL L
Sbjct: 533 FLFVILLFLLLIL---FLFLISFIILTFIIFSLFL*CVCFFIFILLFLFLLLLFIFLFLL 363
>TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (42%)
Length = 655
Score = 40.0 bits (92), Expect = 0.007
Identities = 18/46 (39%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Frame = +3
Query: 226 RRGKGHDMQQVQCFSCKQFGHIARSC--SKKFCNYCKQRGHIITEC 269
R G G + V C +C+Q GH++R C C+ C RGH+ EC
Sbjct: 282 RGGGGGGYRDVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYEC 419
Score = 37.0 bits (84), Expect = 0.057
Identities = 14/33 (42%), Positives = 22/33 (66%), Gaps = 1/33 (3%)
Frame = +3
Query: 238 CFSCKQFGHIARSCS-KKFCNYCKQRGHIITEC 269
C +C + GHIA C+ +K CN C++ GH+ +C
Sbjct: 78 CNNCYKQGHIAAECTNEKACNNCRKTGHLARDC 176
Score = 33.5 bits (75), Expect = 0.63
Identities = 15/38 (39%), Positives = 20/38 (52%), Gaps = 8/38 (21%)
Frame = +3
Query: 240 SCKQFGHIARSCSK--------KFCNYCKQRGHIITEC 269
+C + GH AR CS + CN C ++GHI EC
Sbjct: 6 TCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAEC 119
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 40.0 bits (92), Expect = 0.007
Identities = 18/46 (39%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Frame = +2
Query: 226 RRGKGHDMQQVQCFSCKQFGHIARSC--SKKFCNYCKQRGHIITEC 269
R G G + V C +C+Q GH++R C C+ C RGH+ EC
Sbjct: 806 RGGGGGGYRDVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYEC 943
Score = 38.5 bits (88), Expect = 0.019
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Frame = +2
Query: 238 CFSCKQFGHIARSC-SKKFCNYCKQRGHIITECYVRPPP 275
C++CK+ GH+A SC ++ C+ C + GH EC P P
Sbjct: 458 CWNCKEPGHMASSCPNEGICHTCGKAGHRARECSAPPMP 574
Score = 37.0 bits (84), Expect = 0.057
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Frame = +2
Query: 238 CFSCKQFGHIARSC-SKKFCNYCKQRGHIITEC 269
C +C++ GH+AR C + CN C GH+ +C
Sbjct: 650 CNNCRKTGHLARDCPNDPICNLCNVSGHVARQC 748
Score = 37.0 bits (84), Expect = 0.057
Identities = 14/33 (42%), Positives = 22/33 (66%), Gaps = 1/33 (3%)
Frame = +2
Query: 238 CFSCKQFGHIARSCS-KKFCNYCKQRGHIITEC 269
C +C + GHIA C+ +K CN C++ GH+ +C
Sbjct: 593 CNNCYKQGHIAAECTNEKACNNCRKTGHLARDC 691
Score = 36.6 bits (83), Expect = 0.074
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 8/40 (20%)
Frame = +2
Query: 238 CFSCKQFGHIARSCSK--------KFCNYCKQRGHIITEC 269
C +C + GH AR CS + CN C ++GHI EC
Sbjct: 515 CHTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAEC 634
Score = 35.0 bits (79), Expect = 0.22
Identities = 19/45 (42%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Frame = +2
Query: 226 RRGKGHDMQQVQCFSCKQFGHIARSC-SKKFCNYCKQRGHIITEC 269
RRG D C +CK+ GH AR C + C+ C GHI +EC
Sbjct: 317 RRGFSRDNL---CKNCKRPGHYARECPNVAICHNCGLPGHIASEC 442
Score = 33.5 bits (75), Expect = 0.63
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Frame = +2
Query: 238 CFSCKQFGHIARSCS-KKFCNYCKQRGHIITEC 269
C +C GHIA C+ K C CK+ GH+ + C
Sbjct: 401 CHNCGLPGHIASECTTKSLCWNCKEPGHMASSC 499
>BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delalandii},
partial (5%)
Length = 667
Score = 39.3 bits (90), Expect = 0.011
Identities = 31/111 (27%), Positives = 50/111 (44%), Gaps = 12/111 (10%)
Frame = -3
Query: 314 IGIQGKSSNASHPWFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVG--- 370
I + ++ S W+ DSGAS+H+T + + + ++ +I I +G L+I G
Sbjct: 656 IFLTSSAATPSQSWYPDSGASHHVTNMSQNIQQVAPFEEPIQIIIGNGQGLNINSSGLST 477
Query: 371 ---DINSDF----RDVLVSPGLASNLLSVGQLVV--ILMLIFHVLVVLCKS 412
IN F ++L P + NL+ V Q + FH V L KS
Sbjct: 476 FSSPINPQFSLVLSNLLFVPTITKNLIRVSQFCKDNNVYFEFHSYVCLFKS 324
>TC233180 similar to UP|Q944K0 (Q944K0) At1g18030/T10F20_3, partial (11%)
Length = 916
Score = 38.9 bits (89), Expect = 0.015
Identities = 31/95 (32%), Positives = 49/95 (50%), Gaps = 12/95 (12%)
Frame = +2
Query: 313 SIGIQGKSS-----NASHPW---FLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTL 364
S+ + GKSS NAS +DSG + HMT Y + GN+ I +A+G+ +
Sbjct: 386 SLTMAGKSSSFLSFNASGTENI*IIDSGVTDHMTPHSSYFSSYTFLIGNQHIIVANGSHI 565
Query: 365 SITDVGDI----NSDFRDVLVSPGLASNLLSVGQL 395
I G+I + +VL P L++NLLS+ ++
Sbjct: 566 PIIGCGNIQLQSSLHLNNVLYVPKLSNNLLSIHKI 670
>TC211663
Length = 426
Score = 38.1 bits (87), Expect = 0.025
Identities = 28/117 (23%), Positives = 54/117 (45%)
Frame = -3
Query: 83 FSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNLWTEHSAIIHA 142
F A +WN LK ++Q + + +L+ EI KQG+ +V ++++ +W E + +
Sbjct: 403 FDHATNIWNDLKEGFSQGDLLQIAELQEEIYRLKQGSHTVLDFFTKLKFVWEE---LDNY 233
Query: 143 DVPNIYLAAVQEVYNTSKQDQFLMILRPEFEVVRGSLLNRNVVPSLDTCVSELLREE 199
+ N+ + + FL L F VV +L + +PS S +++ E
Sbjct: 232 GLMNLCTCPSRTYHQQDFVIHFLKGLDERFSVVCSEVLLMDHLPSTKRIFSMVIQHE 62
>TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like protein, partial
(62%)
Length = 1131
Score = 36.6 bits (83), Expect = 0.074
Identities = 20/55 (36%), Positives = 25/55 (45%), Gaps = 4/55 (7%)
Frame = +1
Query: 228 GKGHDMQQVQCFSCKQFGHIARSCS----KKFCNYCKQRGHIITECYVRPPPSTQ 278
G+G +CF+C GH AR C K C C +RGHI C P +Q
Sbjct: 334 GRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKKLSQ 498
>TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (69%)
Length = 1623
Score = 36.2 bits (82), Expect = 0.097
Identities = 19/50 (38%), Positives = 23/50 (46%), Gaps = 4/50 (8%)
Frame = +2
Query: 228 GKGHDMQQVQCFSCKQFGHIARSCS----KKFCNYCKQRGHIITECYVRP 273
G+G +CF+C GH AR C K C C +RGHI C P
Sbjct: 284 GRGPPPGSGRCFNCGLDGHWARDCKAGDWKNKCYRCGERGHIERNCKNSP 433
>TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K24M7, partial (36%)
Length = 1224
Score = 34.7 bits (78), Expect = 0.28
Identities = 13/40 (32%), Positives = 22/40 (54%), Gaps = 8/40 (20%)
Frame = +1
Query: 238 CFSCKQFGHIARSCSK--------KFCNYCKQRGHIITEC 269
C C++ GH A++C + K+C C + GH +T+C
Sbjct: 346 CLRCRRRGHRAKNCPEVLDGAKDAKYCYNCGENGHALTQC 465
Score = 33.9 bits (76), Expect = 0.48
Identities = 15/39 (38%), Positives = 18/39 (45%), Gaps = 7/39 (17%)
Frame = +1
Query: 238 CFSCKQFGHIARSC-------SKKFCNYCKQRGHIITEC 269
CF CK HIA+ C K C C++RGH C
Sbjct: 271 CFICKAMDHIAKLCPEKAEWEKNKICLRCRRRGHRAKNC 387
>BG508014 GP|15128578|db prolactin receptor {Paralichthys olivaceus}, partial
(2%)
Length = 450
Score = 33.9 bits (76), Expect = 0.48
Identities = 27/115 (23%), Positives = 47/115 (40%), Gaps = 13/115 (11%)
Frame = +1
Query: 20 NYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALEE-------------WETKDAQIIT 66
NY W Q + +K L L V+ +K AL + W+ +D +
Sbjct: 103 NYLLWLQQVELVIKSRHLHHFL--VNPLIPKKYALVKDCDSDTTTEVYHGWKEQDQFFLA 276
Query: 67 WILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSS 121
W+ STI M+ ++ + ++W+ L+ + A+ QL E+ GN S
Sbjct: 277 WLRSTIYGDMLTHVIGCKSSWQLWDRLQLHFQSLTRARVRQLRNELRCLSHGNRS 441
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 33.9 bits (76), Expect = 0.48
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Frame = +1
Query: 238 CFSCKQFGHIARSCSK-KFCNYCKQRGHIITEC 269
C+ C GH AR CSK + C CK+ GH +C
Sbjct: 127 CYVCGCLGHNARQCSKVQDCFICKKGGHRAKDC 225
Score = 32.3 bits (72), Expect = 1.4
Identities = 18/53 (33%), Positives = 23/53 (42%), Gaps = 13/53 (24%)
Frame = +1
Query: 230 GHDMQQVQ----CFSCKQFGHIARSCSKK---------FCNYCKQRGHIITEC 269
GH+ +Q CF CK+ GH A+ C +K C C GH I C
Sbjct: 148 GHNARQCSKVQDCFICKKGGHRAKDCPEKHTSTSKSIAICLKCGNSGHDIFSC 306
Score = 32.0 bits (71), Expect = 1.8
Identities = 13/46 (28%), Positives = 19/46 (41%), Gaps = 11/46 (23%)
Frame = +1
Query: 235 QVQCFSCKQFGHIARSCSK-----------KFCNYCKQRGHIITEC 269
++ C+ C Q GH +CS+ C C + GH EC
Sbjct: 409 EISCYKCGQLGHTGLACSRLRDEITSGATPSSCFKCGEEGHFAREC 546
Score = 31.2 bits (69), Expect = 3.1
Identities = 15/36 (41%), Positives = 19/36 (52%), Gaps = 4/36 (11%)
Frame = +1
Query: 238 CFSCKQFGHIARSCS----KKFCNYCKQRGHIITEC 269
CF+C + GH A +CS KK C C GH +C
Sbjct: 61 CFNCGEEGHAAVNCSAVKRKKPCYVCGCLGHNARQC 168
Score = 30.4 bits (67), Expect = 5.3
Identities = 13/44 (29%), Positives = 22/44 (49%), Gaps = 6/44 (13%)
Frame = +1
Query: 232 DMQQVQCFSCKQFGHIA------RSCSKKFCNYCKQRGHIITEC 269
D++++QC+ CK+ GH+ + + C C Q GH C
Sbjct: 328 DLKEIQCYVCKRLGHLCCVNTDDATPGEISCYKCGQLGHTGLAC 459
>BI784757
Length = 430
Score = 33.5 bits (75), Expect = 0.63
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Frame = +1
Query: 555 FLYLRNF*HMLKLNFKQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCP 604
F F +M K ++K T+ GGEY+S EFQE+ +GI+ + + P
Sbjct: 277 FEVFEKFKNMAKKQSGSLIKILRTNGGGEYVSTEFQEFCDQQGIIHEVTPP 429
>CO981879
Length = 576
Score = 32.0 bits (71), Expect = 1.8
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Frame = -1
Query: 559 RNF*HMLKLNFKQVLKFF-TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
+ F M++ F+ +K F +D+G EY + + GI+ Q SC +TP
Sbjct: 564 KTFFQMIQTQFQVKIKVFRSDNGREYFNKHLSKXXLENGIIHQSSCVDTP 415
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.347 0.154 0.508
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 68,188,894
Number of Sequences: 63676
Number of extensions: 1168653
Number of successful extensions: 14326
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 10445
Number of HSP's successfully gapped in prelim test: 475
Number of HSP's that attempted gapping in prelim test: 3108
Number of HSP's gapped (non-prelim): 11842
length of query: 1309
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1200
effective length of database: 5,698,948
effective search space: 6838737600
effective search space used: 6838737600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146758.11


