

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.7 - phase: 0
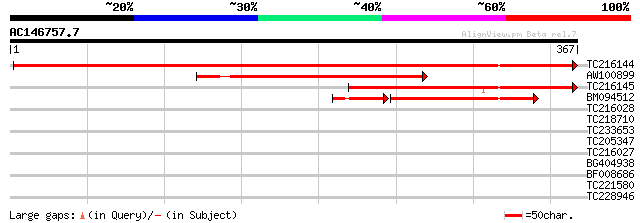
(367 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216144 UP|Q6PP99 (Q6PP99) Mitochondrial pyruvate dehydrogenase... 653 0.0
AW100899 similar to GP|4049632|gb| pyruvate dehydrogenase kinase... 246 9e-66
TC216145 similar to UP|Q6PP98 (Q6PP98) Mitochondrial pyruvate de... 245 2e-65
BM094512 homologue to GP|4049632|gb| pyruvate dehydrogenase kina... 175 9e-51
TC216028 homologue to UP|Q9ZSQ0 (Q9ZSQ0) Ethylene response senso... 32 0.35
TC218710 similar to UP|Q949H7 (Q949H7) Ethylene receptor, partia... 31 0.77
TC233653 similar to UP|ETR1_MALDO (O81122) Ethylene receptor , ... 30 1.3
TC205347 30 2.2
TC216027 homologue to UP|Q9ZSQ0 (Q9ZSQ0) Ethylene response senso... 29 3.8
BG404938 28 5.0
BF008686 28 6.5
TC221580 28 6.5
TC228946 28 6.5
>TC216144 UP|Q6PP99 (Q6PP99) Mitochondrial pyruvate dehydrogenase kinase
isoform 1 , complete
Length = 1536
Score = 653 bits (1685), Expect = 0.0
Identities = 315/365 (86%), Positives = 350/365 (95%)
Frame = +2
Query: 3 AIEAFSKSLLQEVHKWGCLKQTGVSLRYMMEFGSQPTDKNLLISAQFLQKELAIRIARRA 62
A E FSKSL++EV++WGCLKQTGVSLRYMMEFGS+PT+KNLLISAQFL KELAIRIARRA
Sbjct: 197 ACETFSKSLIEEVNRWGCLKQTGVSLRYMMEFGSKPTNKNLLISAQFLHKELAIRIARRA 376
Query: 63 IELESLPYGLSKKPAILKVRDWYVDSFRDIRSCPEVKDMKDEREFTDVIKAIKVRHNNVV 122
+ELE+LPYGLS+KPA+LKVRDWYVDSFRD+R+ P++K++ DEREFT++IKAIKVRHNNVV
Sbjct: 377 VELENLPYGLSQKPAVLKVRDWYVDSFRDVRAFPDIKNVNDEREFTEMIKAIKVRHNNVV 556
Query: 123 PTMALGVQQLKKELKTKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHVV 182
PTMA+GVQQLKK + KI EDLVEIH+FLDRFY+SRIG+RMLIGQHVELHNPNPPPHVV
Sbjct: 557 PTMAMGVQQLKKGMDPKIVYEDLVEIHQFLDRFYMSRIGIRMLIGQHVELHNPNPPPHVV 736
Query: 183 GYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELVK 242
GYIHTKMSPV VARNASEDARSIC REYGSAP+++IYGDP+FTFPYVPAHLHLMVFELVK
Sbjct: 737 GYIHTKMSPVEVARNASEDARSICCREYGSAPDVHIYGDPNFTFPYVPAHLHLMVFELVK 916
Query: 243 NSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARN 302
NSLRAVQER+M+SDKV+PPIRIIVADG+EDVTIK+SDEGGGI RSGLPKIFTYLYSTARN
Sbjct: 917 NSLRAVQERFMNSDKVAPPIRIIVADGIEDVTIKVSDEGGGIARSGLPKIFTYLYSTARN 1096
Query: 303 PLDEHADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS 362
PLDEH+DLG+ D+V TMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS
Sbjct: 1097PLDEHSDLGIGDNV-TMAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHLSRLGDS 1273
Query: 363 QEPLP 367
QEPLP
Sbjct: 1274QEPLP 1288
>AW100899 similar to GP|4049632|gb| pyruvate dehydrogenase kinase
{Arabidopsis thaliana}, partial (34%)
Length = 430
Score = 246 bits (629), Expect = 9e-66
Identities = 121/149 (81%), Positives = 132/149 (88%)
Frame = +1
Query: 122 VPTMALGVQQLKKELKTKIDSEDLVEIHEFLDRFYLSRIGVRMLIGQHVELHNPNPPPHV 181
VPTMALGVQQLK ED EI EFLDRFY+SRIG+RMLIGQHVELHNPNPPP+
Sbjct: 1 VPTMALGVQQLKNVF------EDPDEIDEFLDRFYMSRIGIRMLIGQHVELHNPNPPPNC 162
Query: 182 VGYIHTKMSPVSVARNASEDARSICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELV 241
VGYIHT MSPV+VARNASEDARS+C REYGSA E+ IYGDPDFTFPYVPAHLHLMVFELV
Sbjct: 163 VGYIHTNMSPVNVARNASEDARSMCYREYGSAAEVRIYGDPDFTFPYVPAHLHLMVFELV 342
Query: 242 KNSLRAVQERYMDSDKVSPPIRIIVADGL 270
KNSLRAVQER+MDSD+V+PPIRII+ADG+
Sbjct: 343 KNSLRAVQERFMDSDEVAPPIRIIIADGI 429
>TC216145 similar to UP|Q6PP98 (Q6PP98) Mitochondrial pyruvate dehydrogenase
kinase isoform 2 , partial (39%)
Length = 685
Score = 245 bits (626), Expect = 2e-65
Identities = 121/151 (80%), Positives = 134/151 (88%), Gaps = 3/151 (1%)
Frame = +1
Query: 220 GDPDFTFPYVPAHLHLMVFELVKNSLRAVQERYMDSDKVSPPIRIIVADGLEDVTIKISD 279
GD DFTF YVP HLHLMVFELVKNSLRAVQER+MDSD+V+PPIRII+ADG+EDVTIK+ D
Sbjct: 1 GDXDFTFXYVPXHLHLMVFELVKNSLRAVQERFMDSDEVAPPIRIIIADGIEDVTIKVXD 180
Query: 280 EGGGIPRSGLPKIFTYLYSTARNPLD---EHADLGVADSVTTMAGYGYGLPISRLYARYF 336
EGGGIPRSGLP+IFTY STA+N E +D+G ++V TMAGYGYGLPI RLYARYF
Sbjct: 181 EGGGIPRSGLPRIFTYXXSTAKNSSSVEHEPSDIGTMENV-TMAGYGYGLPICRLYARYF 357
Query: 337 GGDLQIISMEGYGTDAYLHLSRLGDSQEPLP 367
GGDLQ+ISMEGYGTDAYLHLSRLGDSQEPLP
Sbjct: 358 GGDLQVISMEGYGTDAYLHLSRLGDSQEPLP 450
>BM094512 homologue to GP|4049632|gb| pyruvate dehydrogenase kinase
{Arabidopsis thaliana}, partial (30%)
Length = 426
Score = 175 bits (444), Expect(2) = 9e-51
Identities = 85/96 (88%), Positives = 93/96 (96%)
Frame = +1
Query: 247 AVQERYMDSDKVSPPIRIIVADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDE 306
AVQER+M+SDKV+PPIRIIVADG+EDVTIK+SDEGGGI RSGLPKIFTYLYSTARNPLDE
Sbjct: 142 AVQERFMNSDKVAPPIRIIVADGIEDVTIKVSDEGGGIARSGLPKIFTYLYSTARNPLDE 321
Query: 307 HADLGVADSVTTMAGYGYGLPISRLYARYFGGDLQI 342
H+DLG+ D+V TMAGYGYGLPISRLYARYFGGDLQI
Sbjct: 322 HSDLGIGDNV-TMAGYGYGLPISRLYARYFGGDLQI 426
Score = 43.1 bits (100), Expect(2) = 9e-51
Identities = 23/36 (63%), Positives = 26/36 (71%)
Frame = +2
Query: 210 YGSAPEINIYGDPDFTFPYVPAHLHLMVFELVKNSL 245
+ S P +N +P F YVPAHLHLMVFELVKNSL
Sbjct: 38 FHSNPRLN--PNPLFLSLYVPAHLHLMVFELVKNSL 139
>TC216028 homologue to UP|Q9ZSQ0 (Q9ZSQ0) Ethylene response sensor, partial
(78%)
Length = 1745
Score = 32.3 bits (72), Expect = 0.35
Identities = 20/78 (25%), Positives = 32/78 (40%)
Frame = +2
Query: 267 ADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGVADSVTTMAGYGYGL 326
+DG + +++ D G GIP +P +FT + P +G G GL
Sbjct: 1088 SDGHFYIRVQVKDSGCGIPPQEIPHLFTKFAQSRSGP------------ARPSSGAGLGL 1231
Query: 327 PISRLYARYFGGDLQIIS 344
I + + GG + I S
Sbjct: 1232 AICKRFVNLMGGHIWIES 1285
>TC218710 similar to UP|Q949H7 (Q949H7) Ethylene receptor, partial (32%)
Length = 1094
Score = 31.2 bits (69), Expect = 0.77
Identities = 25/79 (31%), Positives = 32/79 (39%), Gaps = 4/79 (5%)
Frame = +1
Query: 275 IKISDEGGGIPRSGLPKIFTYLYS----TARNPLDEHADLGVADSVTTMAGYGYGLPISR 330
+++ D G GI +PKIFT T RNP AG G GL I R
Sbjct: 34 VQVKDSGSGINPLDIPKIFTKFAQNQSLTTRNP----------------AGSGLGLAICR 165
Query: 331 LYARYFGGDLQIISMEGYG 349
+ G + + S EG G
Sbjct: 166 RFVNLMEGHIWVES-EGIG 219
>TC233653 similar to UP|ETR1_MALDO (O81122) Ethylene receptor , partial
(29%)
Length = 663
Score = 30.4 bits (67), Expect = 1.3
Identities = 27/83 (32%), Positives = 34/83 (40%), Gaps = 3/83 (3%)
Frame = +1
Query: 275 IKISDEGGGIPRSGLPKIFTYL---YSTARNPLDEHADLGVADSVTTMAGYGYGLPISRL 331
+++ D G GI +PK+FT S RNP AG G GL I R
Sbjct: 484 VQVKDSGSGINPQDIPKLFTKFAQNQSLTRNP----------------AGSGLGLXICRR 615
Query: 332 YARYFGGDLQIISMEGYGTDAYL 354
Y I+ + YGT AYL
Sbjct: 616 Y--------YILVRKSYGT-AYL 657
>TC205347
Length = 1514
Score = 29.6 bits (65), Expect = 2.2
Identities = 11/26 (42%), Positives = 18/26 (68%)
Frame = -3
Query: 170 VELHNPNPPPHVVGYIHTKMSPVSVA 195
V L +P PPPH +G+I + P+S++
Sbjct: 858 VPLPSPPPPPHFLGHIKKFLDPISLS 781
>TC216027 homologue to UP|Q9ZSQ0 (Q9ZSQ0) Ethylene response sensor, partial
(30%)
Length = 869
Score = 28.9 bits (63), Expect = 3.8
Identities = 19/78 (24%), Positives = 31/78 (39%)
Frame = +3
Query: 267 ADGLEDVTIKISDEGGGIPRSGLPKIFTYLYSTARNPLDEHADLGVADSVTTMAGYGYGL 326
+DG + +++ D G GI +P +FT + P +G G GL
Sbjct: 177 SDGHFYIRVQVKDSGCGILPQDIPHLFTKFAQSRSGP------------ARPSSGAGLGL 320
Query: 327 PISRLYARYFGGDLQIIS 344
I + + GG + I S
Sbjct: 321 AICKRFVNLMGGHIWIES 374
>BG404938
Length = 403
Score = 28.5 bits (62), Expect = 5.0
Identities = 15/42 (35%), Positives = 19/42 (44%)
Frame = -1
Query: 170 VELHNPNPPPHVVGYIHTKMSPVSVARNASEDARSICMREYG 211
VE PNP PH V + SP + S + R C + YG
Sbjct: 265 VEAQEPNPRPHGVYPLSISRSPCTGRPCVSREGRRKCGQGYG 140
>BF008686
Length = 349
Score = 28.1 bits (61), Expect = 6.5
Identities = 9/21 (42%), Positives = 15/21 (70%)
Frame = -2
Query: 175 PNPPPHVVGYIHTKMSPVSVA 195
P PPPH +G+I + P+S++
Sbjct: 267 PPPPPHFIGHIKKILDPISLS 205
>TC221580
Length = 526
Score = 28.1 bits (61), Expect = 6.5
Identities = 10/29 (34%), Positives = 18/29 (61%)
Frame = +2
Query: 163 RMLIGQHVELHNPNPPPHVVGYIHTKMSP 191
R ++G H L +P+PPPH + + ++P
Sbjct: 128 RQILGHHHFLFSPSPPPHPPPHPSSSLTP 214
>TC228946
Length = 601
Score = 28.1 bits (61), Expect = 6.5
Identities = 9/38 (23%), Positives = 21/38 (54%)
Frame = -1
Query: 205 ICMREYGSAPEINIYGDPDFTFPYVPAHLHLMVFELVK 242
+C + Y P+ +++ DP+ + +VP H M +++
Sbjct: 130 VCQKSYQDTPQTDMWRDPEESCDWVPFHTSSMAASILE 17
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,007,768
Number of Sequences: 63676
Number of extensions: 176768
Number of successful extensions: 974
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 958
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 970
length of query: 367
length of database: 12,639,632
effective HSP length: 99
effective length of query: 268
effective length of database: 6,335,708
effective search space: 1697969744
effective search space used: 1697969744
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146757.7


