

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.12 + phase: 0 /partial
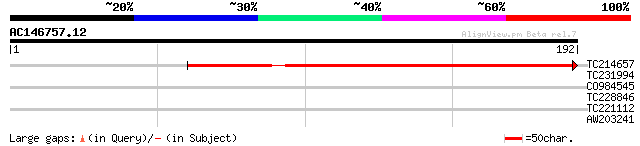
(192 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC214657 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxid... 197 3e-51
TC231994 weakly similar to UP|Q9FLV6 (Q9FLV6) Emb|CAB61996.1, pa... 36 0.010
CO984545 36 0.013
TC228846 similar to UP|Q9LMV4 (Q9LMV4) F5M15.23, partial (6%) 30 0.69
TC221112 weakly similar to GB|AAA30742.1|163635|BOVPROTA protami... 27 4.5
AW203241 27 4.5
>TC214657 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxidase precursor
, partial (72%)
Length = 1750
Score = 197 bits (500), Expect = 3e-51
Identities = 96/132 (72%), Positives = 111/132 (83%)
Frame = -3
Query: 61 RSCTFIDGHGGTFDVNIEDLGTSKADLLAPFSAKLIDGINQSEARRRALVLLCFVYMNTN 120
RSCTF+DG GGTFDV++EDL TS+AD LA K++DGINQSEARR AL+L CFVY + N
Sbjct: 1547 RSCTFVDGLGGTFDVDVEDLETSRADPLA----KIVDGINQSEARRTALMLFCFVYKDAN 1380
Query: 121 AKDAYVTSVDRKGFDVLAKVTGPVSKDGVGQYQWKELRFMFEQEANDVETFCQHLVQMEE 180
AKDAY+TS+DRKGFDVLAKV+ PV KDG+ YQWKE RFMF++EANDVE FC LV+MEE
Sbjct: 1379 AKDAYITSIDRKGFDVLAKVSSPVLKDGIDGYQWKEFRFMFKEEANDVEMFCSQLVEMEE 1200
Query: 181 EVIKKVSASSGL 192
EVI KVS SSGL
Sbjct: 1199 EVINKVSTSSGL 1164
>TC231994 weakly similar to UP|Q9FLV6 (Q9FLV6) Emb|CAB61996.1, partial (29%)
Length = 891
Score = 36.2 bits (82), Expect = 0.010
Identities = 24/107 (22%), Positives = 47/107 (43%), Gaps = 1/107 (0%)
Frame = +1
Query: 31 SSGGSSTSRSDNLQKLLEVTEKYSVYRFKTRSCTFIDGHGGTFDVNIEDLGTSKADLLAP 90
SSG ++ + +N + + + + S Y+ + G + +ED ++ D++A
Sbjct: 31 SSGDNAAQQVENNENSVXXSCETSFYKLEMIKIQVFSAQGQPTALELEDYMNAQPDIIAH 210
Query: 91 FSAKLIDGIN-QSEARRRALVLLCFVYMNTNAKDAYVTSVDRKGFDV 136
K+I + E AL LC+ ++A + +D GFDV
Sbjct: 211 SXXKIISRLKADGEKTLEALKSLCWRCKGIQVEEAQLICLDSLGFDV 351
>CO984545
Length = 798
Score = 35.8 bits (81), Expect = 0.013
Identities = 24/107 (22%), Positives = 49/107 (45%), Gaps = 1/107 (0%)
Frame = -2
Query: 31 SSGGSSTSRSDNLQKLLEVTEKYSVYRFKTRSCTFIDGHGGTFDVNIEDLGTSKADLLAP 90
SSG ++ + +N + + + + S Y+ + G + +ED ++ D++A
Sbjct: 662 SSGDNAAQQVENNENSVIPSCETSFYKLEMIKIQVFSAQGQPTALELEDYMNAQPDIIAH 483
Query: 91 FSAKLIDGIN-QSEARRRALVLLCFVYMNTNAKDAYVTSVDRKGFDV 136
++K+I + E AL LC+ ++A + +D GFDV
Sbjct: 482 SASKIISRLKADGEKTLEALKSLCWRCKGIQVEEAQLICLDSLGFDV 342
>TC228846 similar to UP|Q9LMV4 (Q9LMV4) F5M15.23, partial (6%)
Length = 645
Score = 30.0 bits (66), Expect = 0.69
Identities = 19/52 (36%), Positives = 29/52 (55%), Gaps = 3/52 (5%)
Frame = +2
Query: 12 IRSEEKATRKFSYTVSG---VLSSGGSSTSRSDNLQKLLEVTEKYSVYRFKT 60
+RS + K + T SG VLS S+TSR+ + +KLL++ S+ F T
Sbjct: 284 VRSTKSMASKKTSTPSGSTTVLSPTASTTSRTTSFEKLLKIPPASSLLAFAT 439
>TC221112 weakly similar to GB|AAA30742.1|163635|BOVPROTA protamine {Bos
taurus;} , partial (43%)
Length = 814
Score = 27.3 bits (59), Expect = 4.5
Identities = 16/46 (34%), Positives = 24/46 (51%), Gaps = 10/46 (21%)
Frame = -1
Query: 121 AKDAYVTSVDRKGFDVLAKV----------TGPVSKDGVGQYQWKE 156
AK VT+ R+G + + V GP +K+G+G Y+WKE
Sbjct: 373 AKAIPVTTATRRGRTLCSFVETTP*RATFWAGPSTKNGLGIYKWKE 236
>AW203241
Length = 401
Score = 27.3 bits (59), Expect = 4.5
Identities = 20/66 (30%), Positives = 26/66 (39%)
Frame = +3
Query: 4 LTEKVHEVIRSEEKATRKFSYTVSGVLSSGGSSTSRSDNLQKLLEVTEKYSVYRFKTRSC 63
LT +VIR + KAT KF L + G + L S Y F +SC
Sbjct: 192 LTNNRIDVIRRKRKATEKFLKKDIADLLANGLDDRAYGRAEGLFVELTLSSCYDFVEQSC 371
Query: 64 TFIDGH 69
F+ H
Sbjct: 372 DFVLKH 389
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,872,972
Number of Sequences: 63676
Number of extensions: 71896
Number of successful extensions: 304
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 303
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 303
length of query: 192
length of database: 12,639,632
effective HSP length: 92
effective length of query: 100
effective length of database: 6,781,440
effective search space: 678144000
effective search space used: 678144000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146757.12


