

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146755.5 - phase: 0
(485 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
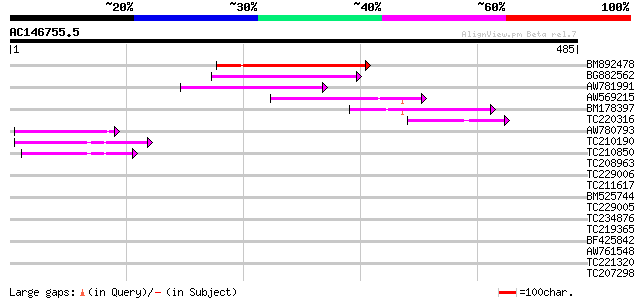
Score E
Sequences producing significant alignments: (bits) Value
BM892478 117 2e-26
BG882562 103 2e-22
AW781991 100 2e-21
AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis... 74 2e-13
BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thalia... 62 6e-10
TC220316 59 5e-09
AW780793 54 2e-07
TC210190 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, part... 52 5e-07
TC210850 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, part... 50 3e-06
TC208963 41 0.001
TC229006 41 0.001
TC211617 39 0.004
BM525744 37 0.020
TC229005 33 0.29
TC234876 similar to UP|Q7RNR5 (Q7RNR5) 60S ribosomal protein L27... 32 0.64
TC219365 similar to UP|Q6IEM2 (Q6IEM2) WRKY transcription factor... 31 1.1
BF425842 31 1.1
AW761548 31 1.1
TC221320 similar to UP|Q6IEM2 (Q6IEM2) WRKY transcription factor... 30 3.2
TC207298 30 3.2
>BM892478
Length = 429
Score = 117 bits (292), Expect = 2e-26
Identities = 53/131 (40%), Positives = 82/131 (62%)
Frame = +3
Query: 178 VNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNT 237
+ + + ++D F+ V +G+ ++FW DG S S FGDVL D +Y+K Y
Sbjct: 33 LEYFQSRQAEDTGFFYAMEVD-NGNCMNIFWADGRSRYSCSHFGDVLVLDTSYRKTVYLV 209
Query: 238 PLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNA 297
P F GVNHH Q ++ G A+I DE+E+++ WL + ++ AM G+LP++VI D D++++ A
Sbjct: 210 PFATFVGVNHHKQPVLLGCALIADESEESFTWLFQTWLRAMSGRLPLTVIADQDIAIQRA 389
Query: 298 IRKVFPEAHHR 308
I KVFP HHR
Sbjct: 390 IAKVFPVTHHR 422
>BG882562
Length = 416
Score = 103 bits (256), Expect = 2e-22
Identities = 49/129 (37%), Positives = 78/129 (59%)
Frame = +3
Query: 173 DAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKK 232
D E N+ + + F+ ++ DG L+++FW D S YS FGDV+AFD+T
Sbjct: 30 DPELISNYFCRIQLMNPNFFYVMDLNDDGQLRNVFWIDSRSRAAYSYFGDVVAFDSTCLS 209
Query: 233 IKYNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDL 292
Y PLV F GVNHH +S++ G ++ DET +TY+WL + ++ M G+ P ++IT+
Sbjct: 210 NNYEIPLVAFVGVNHHGKSVLLGCGLLADETFETYIWLFRAWLTCMTGRPPQTIITNXCK 389
Query: 293 SMRNAIRKV 301
+M++AI +V
Sbjct: 390 AMQSAIAEV 416
>AW781991
Length = 419
Score = 99.8 bits (247), Expect = 2e-21
Identities = 45/127 (35%), Positives = 73/127 (57%), Gaps = 1/127 (0%)
Frame = +3
Query: 147 GGYQNVGYNRRDMYNEQRKRRMR-WNSDAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQH 205
GG VG+ D N R R+R D + +++L+ M +++ F+ D S+ +
Sbjct: 33 GGISKVGFTEVDCRNYMRNNRLRSLEGDIQLVLDYLRQMHAENPNFFYAVQGDEDQSITN 212
Query: 206 LFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVIFSGVNHHNQSIIFGSAIIGDETED 265
+FW D + M+Y+ FGD + FD TY+ +Y P F+GVNHH Q ++FG A + +E+E
Sbjct: 213 VFWADPKARMNYTFFGDTVTFDTTYRSNRYRLPFAFFTGVNHHGQPVLFGCAFLINESEA 392
Query: 266 TYVWLLK 272
++VWL K
Sbjct: 393 SFVWLFK 413
>AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana},
partial (19%)
Length = 425
Score = 73.6 bits (179), Expect = 2e-13
Identities = 39/139 (28%), Positives = 74/139 (53%), Gaps = 6/139 (4%)
Frame = +1
Query: 224 LAFDATYKKIKYNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLP 283
+ FD TY+ Y+ L I+ GV+++ + F A++ DE ++ W LK F+ M GK P
Sbjct: 10 VVFDTTYRVEAYDMLLGIWLGVDNNGMTCFFSCALLRDENIQSFSWALKAFLGFMKGKAP 189
Query: 284 VSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIKNLHFVSKFKDC------LLG 337
+++TD ++ ++ AI P+ H C WH++ + S+ +L S++ + L
Sbjct: 190 QTILTDHNMWLKEAIAVELPQTKHAFCIWHIL-SKFSDWFSLLLGSQYDEWKAEFHRLYN 366
Query: 338 DVDVDVFQRKWEELVTEFG 356
V+ F+ W ++V ++G
Sbjct: 367 LEQVEDFEEGWRQMVDQYG 423
>BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana}, partial
(20%)
Length = 422
Score = 62.0 bits (149), Expect = 6e-10
Identities = 34/131 (25%), Positives = 58/131 (43%), Gaps = 6/131 (4%)
Frame = +1
Query: 291 DLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIKNLHFVSKFKDC------LLGDVDVDVF 344
++ ++ A+ P H C W ++ S N ++ D L V+ F
Sbjct: 4 NICLKEALSTEMPTTKHAFCIWMIVAKFPSWF-NAVLGERYNDWKAEFYRLYNLESVEDF 180
Query: 345 QRKWEELVTEFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVS 404
+ W E+ FGL N M+ +Y R +WA R F AG TT + + +++ +++S
Sbjct: 181 ELGWREMACSFGLHSNRHMVNLYSSRSLWALPFLRSHFLAGMTTTGQSKSINAFIQRFLS 360
Query: 405 ALTNLHDFFQQ 415
A T L F +Q
Sbjct: 361 AQTRLAHFVEQ 393
>TC220316
Length = 988
Score = 58.9 bits (141), Expect = 5e-09
Identities = 31/87 (35%), Positives = 44/87 (49%)
Frame = +1
Query: 341 VDVFQRKWEELVTEFGLEENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFG 400
VD W ++GL+++ W+ EMYQKR W + +G FFAG E L S FG
Sbjct: 25 VDEXXATWNXXXNKYGLKDDAWLKEMYQKRASWVPLYLKGTFFAGIPMN---ESLDSFFG 195
Query: 401 KYVSALTNLHDFFQQFFRWLNYMRYRE 427
++A T L +F ++ R L R E
Sbjct: 196 ALLNAQTPLMEFIPRYERGLERRREEE 276
>AW780793
Length = 419
Score = 53.5 bits (127), Expect = 2e-07
Identities = 32/91 (35%), Positives = 47/91 (51%), Gaps = 1/91 (1%)
Frame = +3
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRN-INGEVVQQTFLCHREGIREEKYINSTSRK 63
F RE A FY YG GF R R+ +N +V+ Q F+C +EG R +KY++ R
Sbjct: 150 FNSREEAREFYIAYGRRIGFTVRIHHNRRSRVNNQVIGQDFVCSKEGFRAKKYLHRKDRV 329
Query: 64 REHKPLSRCGCQAKVRVHIDVSSQRWYIKLF 94
P +R GCQA +R+ + +W + F
Sbjct: 330 LPPPPATREGCQAMIRLALR-DRGKWVVTKF 419
>TC210190 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, partial (65%)
Length = 866
Score = 52.4 bits (124), Expect = 5e-07
Identities = 36/119 (30%), Positives = 54/119 (45%), Gaps = 1/119 (0%)
Frame = +1
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRNI-NGEVVQQTFLCHREGIREEKYINSTSRK 63
F A FYN Y GF R S+L R+ +G + +T +C++EG R R+
Sbjct: 7 FGSEAAAHAFYNAYATEVGFIVRVSKLSRSRRDGTAIGRTLVCNKEGFRMADKREKIVRQ 186
Query: 64 REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSEYDK 122
R +R GC+A + V +SS +W + F +H H K R + +E DK
Sbjct: 187 RAE---TRVGCRAMIMVR-KLSSGKWVVAKFVKEHTHPLTPGKGRRDFYYEQYPTEQDK 351
>TC210850 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, partial (63%)
Length = 1326
Score = 49.7 bits (117), Expect = 3e-06
Identities = 33/100 (33%), Positives = 48/100 (48%), Gaps = 1/100 (1%)
Frame = +1
Query: 11 AFMFYNWYGCFHGFAARKSRLIRNI-NGEVVQQTFLCHREGIREEKYINSTSRKREHKPL 69
A FYN Y GF R S+L R+ +G + +T +C++EG R R+R
Sbjct: 7 AHAFYNAYATDVGFIVRVSKLSRSRRDGTAIGRTLVCNKEGFRMADKREKIVRQRAE--- 177
Query: 70 SRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFER 109
+R GC+A + V +SS +W I F +H H K R
Sbjct: 178 TRVGCRAMIMVR-KLSSGKWVIAKFVKEHTHPLTPGKGRR 294
>TC208963
Length = 1163
Score = 40.8 bits (94), Expect = 0.001
Identities = 19/79 (24%), Positives = 43/79 (54%)
Frame = +1
Query: 282 LPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIKNLHFVSKFKDCLLGDVDV 341
+P ++T GD+++ +A++ VFP + + LC +H+ +N + K++ + + ++ ++ DV
Sbjct: 19 MPQVIVTVGDIALMSAVQVVFPSSSNLLCRFHINQNVKAKCKSIVHLKEKQELMMDAWDV 198
Query: 342 DVFQRKWEELVTEFGLEEN 360
V E + EN
Sbjct: 199 VVNSPNEGEYMQRLAFFEN 255
>TC229006
Length = 991
Score = 40.8 bits (94), Expect = 0.001
Identities = 36/123 (29%), Positives = 54/123 (43%), Gaps = 5/123 (4%)
Frame = +2
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIR-NINGEVVQQTFLCHREGIREEKYINSTSRK 63
F + A +FY+ Y GF R R +G ++ + C++EG Y S K
Sbjct: 119 FESEDAAKIFYDEYARRLGFVMRVMSCRRPERDGRILARRLGCNKEG-----YCVSIRGK 283
Query: 64 ----REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSE 119
R+ + +R GC+A + + + S +W I F DHNH V E A + M E
Sbjct: 284 FASVRKPRASTREGCKAMIHIKYNKSG-KWVITKFVKDHNHPLVVSPRE----ARQTMDE 448
Query: 120 YDK 122
DK
Sbjct: 449 KDK 457
>TC211617
Length = 534
Score = 39.3 bits (90), Expect = 0.004
Identities = 37/171 (21%), Positives = 68/171 (39%), Gaps = 7/171 (4%)
Frame = -1
Query: 46 CHREG---IREEKYINSTSRKREHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSF 102
C R G R+++++ + R +CGC K+R + W +KL HNH
Sbjct: 531 CERSGKYKCRKKEFVRRDTCTR------KCGCPFKIRGKPMHGGEGWTVKLICGIHNHEL 370
Query: 103 VKEKFERMLPAHRKMSEYDKYQMNTMRQSGISTTRIH----GYFASQAGGYQNVGYNRRD 158
K E P ++++ +K + M +S + I + + + + YN R
Sbjct: 369 AKTLVEH--PYAGRLTDDEKNIIADMTKSNVKPRNIQLTLKEHNTNSCTTIKQI-YNARS 199
Query: 159 MYNEQRKRRMRWNSDAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWC 209
Y+ + D E + L + +D + W H + ++ LFWC
Sbjct: 198 AYHSSIR-----GDDTE--MQHLMRLLKRDQYIHW-HRLKDQDVVRDLFWC 70
>BM525744
Length = 411
Score = 37.0 bits (84), Expect = 0.020
Identities = 24/92 (26%), Positives = 38/92 (41%)
Frame = +3
Query: 154 YNRRDMYNEQRKRRMRWNSDAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVS 213
YN R Y +K ++ + + L+H D + W V +++ +FW +
Sbjct: 99 YNARQAYRSSQKGP---RTEMQHLLKLLEH----DQYVCWSRKVDDSDAIRDIFWAHPDA 257
Query: 214 CMDYSIFGDVLAFDATYKKIKYNTPLVIFSGV 245
F VL D TYK +Y PL+ GV
Sbjct: 258 IKLLGSFHTVLFLDNTYKVNRYQLPLLEIVGV 353
>TC229005
Length = 679
Score = 33.1 bits (74), Expect = 0.29
Identities = 20/53 (37%), Positives = 26/53 (48%)
Frame = +1
Query: 70 SRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSEYDK 122
+R GC+A + + D S +W I F DHNH V E A + M E DK
Sbjct: 7 TREGCKAMIHIKYDKSG-KWVIAKFVKDHNHPLVVSPRE----ARQTMDEKDK 150
>TC234876 similar to UP|Q7RNR5 (Q7RNR5) 60S ribosomal protein L27 homolog,
partial (15%)
Length = 920
Score = 32.0 bits (71), Expect = 0.64
Identities = 12/34 (35%), Positives = 19/34 (55%)
Frame = -3
Query: 71 RCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVK 104
+CGC K++V + + W KL + HNH+ K
Sbjct: 597 KCGCLFKLQVKPVLGGEGWMKKLIRESHNHALAK 496
>TC219365 similar to UP|Q6IEM2 (Q6IEM2) WRKY transcription factor 59, partial
(33%)
Length = 788
Score = 31.2 bits (69), Expect = 1.1
Identities = 15/59 (25%), Positives = 24/59 (40%)
Frame = +1
Query: 54 EKYINSTSRKREHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLP 112
+K + S+ R + S GC K RV D + + +D HNH + +P
Sbjct: 427 KKTVKSSPNPRNYYKCSGEGCDVKKRVERDRDDSNYVLTTYDGVHNHQTPSTAYYSQMP 603
>BF425842
Length = 414
Score = 31.2 bits (69), Expect = 1.1
Identities = 21/105 (20%), Positives = 42/105 (40%), Gaps = 5/105 (4%)
Frame = +3
Query: 227 DATYKKIKYNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSV 286
D T+ KYN L+ G + + AI+ ET++ ++W L + + + +
Sbjct: 96 DGTFLTGKYNDTLLTAIGQDGSRDNFPLAFAIVESETKEAWIWFLHYL*RYVTPQPNLCI 275
Query: 287 ITDGDLSMRNAIRKVF-----PEAHHRLCAWHLIRNATSNIKNLH 326
I+D + ++ P+ C H+ N KN++
Sbjct: 276 ISDRGTGLLATLQSKHVGWNGPDVSSVYCIRHIASNFNK*FKNVN 410
>AW761548
Length = 322
Score = 31.2 bits (69), Expect = 1.1
Identities = 19/68 (27%), Positives = 34/68 (49%)
Frame = +2
Query: 36 NGEVVQQTFLCHREGIREEKYINSTSRKREHKPLSRCGCQAKVRVHIDVSSQRWYIKLFD 95
+G ++ +T C++ G N +S ++ C C+A + + I +S RW +
Sbjct: 71 DGRILARTLGCNKHGYCVSLRRNISSVRKPMASTREC-CKALIHI-IYNNSVRWVMPELA 244
Query: 96 DDHNHSFV 103
+DHNHS V
Sbjct: 245 NDHNHSSV 268
>TC221320 similar to UP|Q6IEM2 (Q6IEM2) WRKY transcription factor 59, partial
(33%)
Length = 643
Score = 29.6 bits (65), Expect = 3.2
Identities = 14/59 (23%), Positives = 23/59 (38%)
Frame = +1
Query: 54 EKYINSTSRKREHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLP 112
+K + + R + S GC K RV D + + +D HNH + +P
Sbjct: 397 KKTVKNNPNPRNYYKCSGEGCNVKKRVERDRDDSNYVLTTYDGVHNHESPSTAYYSQIP 573
>TC207298
Length = 1011
Score = 29.6 bits (65), Expect = 3.2
Identities = 12/29 (41%), Positives = 18/29 (61%)
Frame = +3
Query: 201 GSLQHLFWCDGVSCMDYSIFGDVLAFDAT 229
G +QH FW +S +D+ FGD++ F T
Sbjct: 339 GHIQHTFWLICLSRVDWWQFGDLVVFQGT 425
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.139 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,749,043
Number of Sequences: 63676
Number of extensions: 334283
Number of successful extensions: 2110
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 2090
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2109
length of query: 485
length of database: 12,639,632
effective HSP length: 101
effective length of query: 384
effective length of database: 6,208,356
effective search space: 2384008704
effective search space used: 2384008704
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146755.5


