

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146755.12 + phase: 0
(57 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
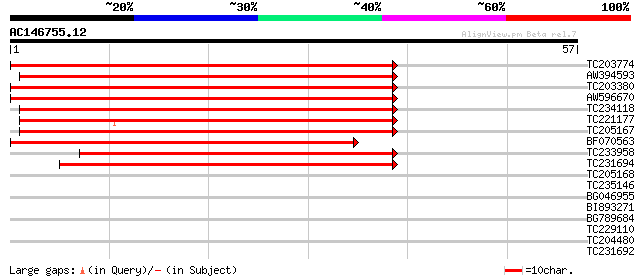
Score E
Sequences producing significant alignments: (bits) Value
TC203774 similar to UP|Q6RVV4 (Q6RVV4) Short-chain dehydrogenase... 65 7e-12
AW394593 62 5e-11
TC203380 similar to UP|Q6RVV4 (Q6RVV4) Short-chain dehydrogenase... 58 9e-10
AW596670 58 9e-10
TC234118 similar to UP|Q6RVV4 (Q6RVV4) Short-chain dehydrogenase... 53 3e-08
TC221177 similar to UP|Q6RVV4 (Q6RVV4) Short-chain dehydrogenase... 52 6e-08
TC205167 similar to UP|Q8L9T6 (Q8L9T6) Putativepod-specific dehy... 50 3e-07
BF070563 47 2e-06
TC233958 weakly similar to UP|Q8LCE7 (Q8LCE7) Putativepod-specif... 44 2e-05
TC231694 weakly similar to UP|Q9W404 (Q9W404) CG3842-PA (Cg3842-... 44 2e-05
TC205168 similar to UP|Q8L9T6 (Q8L9T6) Putativepod-specific dehy... 36 0.004
TC235146 similar to GB|AAM78071.1|21928085|AY125561 AT4g24050/T1... 32 0.052
BG046955 30 0.33
BI893271 26 4.8
BG789684 26 4.8
TC229110 25 6.3
TC204480 similar to UP|Q947C3 (Q947C3) 1-deoxy-D-xylulose-5-phos... 25 6.3
TC231692 similar to UP|Q8W586 (Q8W586) AT4g21800/F17L22_260, par... 25 8.2
>TC203774 similar to UP|Q6RVV4 (Q6RVV4) Short-chain dehydrogenase Tic32,
partial (32%)
Length = 489
Score = 65.1 bits (157), Expect = 7e-12
Identities = 30/39 (76%), Positives = 34/39 (86%)
Frame = +3
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
MWPFS+KG SGFS +STAE+VT GID +GLTAIVTG SS
Sbjct: 171 MWPFSRKGASGFSSSSTAEQVTEGIDGTGLTAIVTGASS 287
>AW394593
Length = 416
Score = 62.4 bits (150), Expect = 5e-11
Identities = 30/38 (78%), Positives = 32/38 (83%)
Frame = +2
Query: 2 WPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
WPFS+ G SGFS STAE+VTHGID SGLTAIVTG SS
Sbjct: 56 WPFSRIGSSGFSSYSTAEEVTHGIDGSGLTAIVTGASS 169
>TC203380 similar to UP|Q6RVV4 (Q6RVV4) Short-chain dehydrogenase Tic32,
partial (97%)
Length = 1209
Score = 58.2 bits (139), Expect = 9e-10
Identities = 27/39 (69%), Positives = 31/39 (79%)
Frame = +2
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
MW F +KG SGFS +STAE+VT GID +G TAIVTG SS
Sbjct: 50 MWLFGRKGASGFSSSSTAEQVTEGIDGTGFTAIVTGASS 166
>AW596670
Length = 299
Score = 58.2 bits (139), Expect = 9e-10
Identities = 27/39 (69%), Positives = 31/39 (79%)
Frame = +2
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
MW F +KG SGFS +STAE+VT GID +G TAIVTG SS
Sbjct: 59 MWLFGRKGASGFSSSSTAEQVTEGIDGTGFTAIVTGASS 175
>TC234118 similar to UP|Q6RVV4 (Q6RVV4) Short-chain dehydrogenase Tic32,
partial (43%)
Length = 479
Score = 53.1 bits (126), Expect = 3e-08
Identities = 25/38 (65%), Positives = 29/38 (75%)
Frame = +3
Query: 2 WPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
WP + G S FS +STAE+VT GID +GLTAIVTG SS
Sbjct: 51 WPCGRNGGSSFSSSSTAEQVTEGIDGTGLTAIVTGASS 164
>TC221177 similar to UP|Q6RVV4 (Q6RVV4) Short-chain dehydrogenase Tic32,
partial (54%)
Length = 709
Score = 52.0 bits (123), Expect = 6e-08
Identities = 27/40 (67%), Positives = 31/40 (77%), Gaps = 2/40 (5%)
Frame = +2
Query: 2 WPFSKKGV--SGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
WPFS+K SGFS +STAE+VT GID + LTAIVTG SS
Sbjct: 11 WPFSRKEKEGSGFSSSSTAEEVTEGIDGTALTAIVTGASS 130
>TC205167 similar to UP|Q8L9T6 (Q8L9T6) Putativepod-specific dehydrogenase
SAC25, partial (93%)
Length = 1242
Score = 49.7 bits (117), Expect = 3e-07
Identities = 24/38 (63%), Positives = 28/38 (73%)
Frame = +1
Query: 2 WPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
W KG SGFS +STAE+VT GID + LTAIVTG +S
Sbjct: 31 WFLGWKGASGFSASSTAEQVTQGIDGTALTAIVTGATS 144
>BF070563
Length = 405
Score = 47.0 bits (110), Expect = 2e-06
Identities = 23/35 (65%), Positives = 26/35 (73%)
Frame = +1
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVT 35
MW KG SGFS +STAE+VT GID + LTAIVT
Sbjct: 7 MWFLGWKGQSGFSASSTAEQVTQGIDGTALTAIVT 111
>TC233958 weakly similar to UP|Q8LCE7 (Q8LCE7) Putativepod-specific
dehydrogenase SAC25, partial (30%)
Length = 365
Score = 43.5 bits (101), Expect = 2e-05
Identities = 22/32 (68%), Positives = 25/32 (77%)
Frame = +2
Query: 8 GVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
G SGF STAE+VT GIDAS LTAI+TG +S
Sbjct: 80 GPSGFGSASTAEQVTDGIDASNLTAIITGGAS 175
>TC231694 weakly similar to UP|Q9W404 (Q9W404) CG3842-PA (Cg3842-pb),
partial (16%)
Length = 498
Score = 43.5 bits (101), Expect = 2e-05
Identities = 21/34 (61%), Positives = 26/34 (75%)
Frame = +3
Query: 6 KKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
+ G SGF +STAE+V GIDAS LTAI+TG +S
Sbjct: 66 RPGCSGFGSSSTAEQVIEGIDASNLTAIITGGAS 167
>TC205168 similar to UP|Q8L9T6 (Q8L9T6) Putativepod-specific dehydrogenase
SAC25, partial (30%)
Length = 512
Score = 36.2 bits (82), Expect = 0.004
Identities = 17/25 (68%), Positives = 21/25 (84%)
Frame = +3
Query: 15 NSTAEKVTHGIDASGLTAIVTGLSS 39
+STAE+VT GID + LTAIVTG +S
Sbjct: 6 SSTAEQVTQGIDGTALTAIVTGATS 80
>TC235146 similar to GB|AAM78071.1|21928085|AY125561 AT4g24050/T19F6_40
{Arabidopsis thaliana;} , partial (49%)
Length = 527
Score = 32.3 bits (72), Expect = 0.052
Identities = 17/33 (51%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Frame = +1
Query: 8 GVSGFSGNSTAEKVTHG-IDASGLTAIVTGLSS 39
G SGF STAE+VT D +TAI+TG +S
Sbjct: 64 GPSGFGSKSTAEQVTENRADLHSITAIITGATS 162
>BG046955
Length = 414
Score = 29.6 bits (65), Expect = 0.33
Identities = 15/30 (50%), Positives = 19/30 (63%), Gaps = 1/30 (3%)
Frame = +3
Query: 8 GVSGFSGNSTAEKVTHG-IDASGLTAIVTG 36
G SGF +TAE+VT D +TAI+TG
Sbjct: 324 GPSGFGSKTTAEQVTENHADLRSITAIITG 413
>BI893271
Length = 429
Score = 25.8 bits (55), Expect = 4.8
Identities = 11/33 (33%), Positives = 17/33 (51%)
Frame = +1
Query: 19 EKVTHGIDASGLTAIVTGLSSITLSYHFFNFKI 51
E +TH I S + G + + YH++N KI
Sbjct: 37 EFLTHTISYSYCLTLFAGFRLLYIIYHYYNIKI 135
>BG789684
Length = 421
Score = 25.8 bits (55), Expect = 4.8
Identities = 13/37 (35%), Positives = 20/37 (53%)
Frame = -1
Query: 3 PFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
PF+K S + S+A HG+ A+ + A+V G S
Sbjct: 334 PFNKPATSAYLSRSSAVVFRHGV-AAAVAAVVVGSPS 227
>TC229110
Length = 956
Score = 25.4 bits (54), Expect = 6.3
Identities = 11/37 (29%), Positives = 17/37 (45%)
Frame = -1
Query: 3 PFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
PF+K S + S+A HG A+ + + G S
Sbjct: 668 PFNKPATSAYLSRSSAVVFRHGFAAAAVVVVAVGSPS 558
>TC204480 similar to UP|Q947C3 (Q947C3) 1-deoxy-D-xylulose-5-phosphate
reductoisomerase, partial (92%)
Length = 2110
Score = 25.4 bits (54), Expect = 6.3
Identities = 18/50 (36%), Positives = 23/50 (46%)
Frame = -2
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSSITLSYHFFNFK 50
M P S +SGFS S ++ I S LTA GL+ T S F+
Sbjct: 765 MIPCSPGMISGFSSTSCKASLSSAIRDSFLTATS*GLNLFTWSARRVTFE 616
>TC231692 similar to UP|Q8W586 (Q8W586) AT4g21800/F17L22_260, partial (19%)
Length = 735
Score = 25.0 bits (53), Expect = 8.2
Identities = 15/36 (41%), Positives = 20/36 (54%), Gaps = 3/36 (8%)
Frame = +3
Query: 15 NSTAEKVTHGIDASGLTAIVTGLSSIT---LSYHFF 47
N+TAEK+T I L + +S T LS+HFF
Sbjct: 105 NNTAEKITSSIGK*TLITVPETPNSTTNTFLSFHFF 212
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,479,842
Number of Sequences: 63676
Number of extensions: 21252
Number of successful extensions: 159
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 159
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 159
length of query: 57
length of database: 12,639,632
effective HSP length: 33
effective length of query: 24
effective length of database: 10,538,324
effective search space: 252919776
effective search space used: 252919776
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146755.12


