

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.6 + phase: 0
(346 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
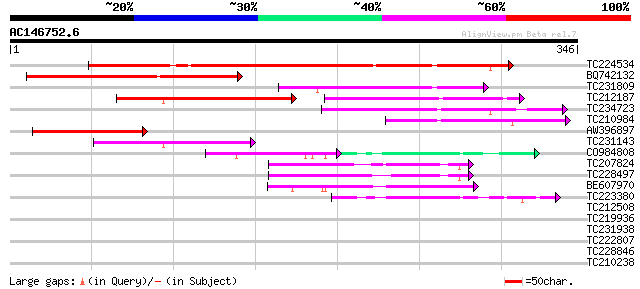
Score E
Sequences producing significant alignments: (bits) Value
TC224534 similar to UP|Q6USA0 (Q6USA0) Transposase (Fragment), p... 198 3e-51
BQ742132 weakly similar to GP|28416671|gb| At5g41980 {Arabidopsi... 120 1e-27
TC231809 112 2e-25
TC212187 weakly similar to UP|Q6USA0 (Q6USA0) Transposase (Fragm... 105 4e-23
TC234723 similar to UP|Q9FHY5 (Q9FHY5) Emb|CAB72466.1 (At5g41980... 92 3e-19
TC210984 70 2e-12
AW396897 similar to GP|28416671|gb| At5g41980 {Arabidopsis thali... 67 2e-11
TC231143 60 2e-09
CO984808 34 5e-06
TC207824 similar to GB|AAN73294.1|25141199|BT002297 At5g12010/F1... 47 1e-05
TC228497 45 4e-05
BE607970 44 1e-04
TC223380 weakly similar to GB|AAP80175.1|32362295|BT009674 At3g5... 41 7e-04
TC212508 39 0.005
TC219936 similar to UP|Q9LJL8 (Q9LJL8) Emb|CAB43653.1 (AT3g19120... 35 0.038
TC231938 similar to UP|Q9LJL8 (Q9LJL8) Emb|CAB43653.1 (AT3g19120... 33 0.15
TC222807 similar to UP|Q9FK34 (Q9FK34) Gb|AAD25781.1, partial (13%) 29 3.6
TC228846 similar to UP|Q9LMV4 (Q9LMV4) F5M15.23, partial (6%) 29 3.6
TC210238 similar to UP|Q9SW27 (Q9SW27) Geranylgeranylated protei... 28 4.7
>TC224534 similar to UP|Q6USA0 (Q6USA0) Transposase (Fragment), partial (50%)
Length = 968
Score = 198 bits (504), Expect = 3e-51
Identities = 118/267 (44%), Positives = 168/267 (62%), Gaps = 8/267 (2%)
Frame = +1
Query: 49 SLYILTHNAKNREVNFWFRRSGETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSST 108
SL + +A + ++ + RS ETIS+ + VL+AI+++ ++++ D + LE S
Sbjct: 61 SLPLSHSHAPHVAISHNYCRSTETISKQIKNVLRAIMKVSKEYLKFHDYN---LEGSVEI 231
Query: 109 RFYPYFKDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEG 168
+ + +FK+ +GA+DG HI V V+A+D PRY RK NVL AC +L+F YVL GWEG
Sbjct: 232 K-WRWFKNSIGALDGIHIPVTVTAEDRPRYHNRKGDIFTNVLGACGPNLRFIYVLPGWEG 408
Query: 169 SASDSRIIKNALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLN 228
SA DSR++ +AL R++ L IP GKY+LVDAG+ G +TPY RYH E+ N P N
Sbjct: 409 SAGDSRVL*DALHRQNCLHIPNGKYFLVDAGYTNGPGFLTPY*RTRYHRNEWIG-NTPQN 585
Query: 229 YKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSA 288
YKELFNLRHAS +N IER+FGVLKKR+ IL T + IK Q II C +L N++
Sbjct: 586 YKELFNLRHASAKNVIERSFGVLKKRWSIL--RTPSFFDIKTQIRIINTCFMLRNFIRDE 759
Query: 289 EPNE--------DLIAEVDAELANQNV 307
+ ++ +L++ VD EL NQ +
Sbjct: 760 QQSDPILEAQDLELLSVVDNELINQQM 840
>BQ742132 weakly similar to GP|28416671|gb| At5g41980 {Arabidopsis thaliana},
partial (25%)
Length = 429
Score = 120 bits (300), Expect = 1e-27
Identities = 62/132 (46%), Positives = 83/132 (61%)
Frame = +3
Query: 11 RNIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSG 70
R ++RM F KLCD+L + LR T +EEQ+A L ++ HN +NR + F+ SG
Sbjct: 33 REMLRMDKHVFHKLCDILRQRAMLRDTAGVMIEEQLAIFLNVIGHNERNRVIQERFQHSG 212
Query: 71 ETISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKV 130
ETISRH + VLKAI L + ++QP P EI ++ RFYPYFKDC+G IDG +I V
Sbjct: 213 ETISRHFNNVLKAIKSLSRE-VLQPPQLKTPPEILNNARFYPYFKDCIGVIDGMNIPAHV 389
Query: 131 SAKDAPRYRGRK 142
AKD R+R +K
Sbjct: 390 PAKDQSRFRNKK 425
>TC231809
Length = 760
Score = 112 bits (280), Expect = 2e-25
Identities = 62/131 (47%), Positives = 78/131 (59%), Gaps = 3/131 (2%)
Frame = +3
Query: 165 GWEGSASDSRIIKNALTREDKL--KIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSA 222
G EGS D ++ L D+ +IP+GKYYLV G++ T G I PY+GVRY EF
Sbjct: 3 GGEGSVXDXXVLXXVLDXXDQXXPQIPQGKYYLVXKGYLNTEGFIAPYQGVRYQHYEFRG 182
Query: 223 RNP-PLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCIL 281
N P N KELFN RH LRNAI R+F VLK RF IL P Y + Q+ I+ A C+L
Sbjct: 183 ANQLPRNAKELFNHRHCFLRNAILRSFNVLKTRFPIL--KLAPQYSFQIQRDIVIAGCVL 356
Query: 282 HNYLMSAEPNE 292
HN++ E N+
Sbjct: 357 HNFIRREERND 389
>TC212187 weakly similar to UP|Q6USA0 (Q6USA0) Transposase (Fragment),
partial (45%)
Length = 973
Score = 105 bits (261), Expect = 4e-23
Identities = 56/112 (50%), Positives = 68/112 (60%), Gaps = 2/112 (1%)
Frame = +2
Query: 66 FRRSGETISRHLHQVLKAILELEEKFI--VQPDGSTIPLEISSSTRFYPYFKDCVGAIDG 123
F S ETI RHLH VL+A+ + I V P EI R+ PYF+DC+GAIDG
Sbjct: 41 F*HSSETIYRHLHSVLEAVCMFAKDIIKPVDPSFRDTLDEILKDARYRPYFRDCIGAIDG 220
Query: 124 THIRVKVSAKDAPRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRI 175
THIRV V + Y GRK Y T NV+ C F + FT+V AGWEGSA D++I
Sbjct: 221 THIRVCVPSHLQGVYSGRKGYTTTNVMIVCDFSMCFTFVWAGWEGSAHDTKI 376
Score = 73.2 bits (178), Expect = 2e-13
Identities = 40/122 (32%), Positives = 66/122 (53%)
Frame = +1
Query: 193 YYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVLK 252
+YLVD+ + G + Y RYHL +F E+FN H+SL++ IERAFG+ K
Sbjct: 514 FYLVDSCYPTFMGFLGSYNKTRYHLPQFRIGPRIRGRVEVFNYYHSSLQSTIERAFGLCK 693
Query: 253 KRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAELANQNVSHDNH 312
R++IL N + + +K Q II AC +HN++ + ++ +D + N+ + D
Sbjct: 694 ARWKILGNMS--PFALKTQNQIIVACMTIHNFIQRNDKSDGEFDSLDED--NEYIDTDKD 861
Query: 313 EA 314
E+
Sbjct: 862 ES 867
>TC234723 similar to UP|Q9FHY5 (Q9FHY5) Emb|CAB72466.1 (At5g41980), partial
(24%)
Length = 837
Score = 92.4 bits (228), Expect = 3e-19
Identities = 52/172 (30%), Positives = 86/172 (49%), Gaps = 22/172 (12%)
Frame = +2
Query: 191 GKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGV 250
GKYY+VD+ + G + PY Y+ KEF + P + ELFN RH+ LR+ I+R FG+
Sbjct: 137 GKYYIVDSKYQNVPGFVAPYSSTPYYSKEFLSDYHPQDASELFNQRHSLLRHVIDRTFGI 316
Query: 251 LKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNE------------------ 292
LK RF IL ++ P+Y ++ Q ++ A C LHNY+ +P++
Sbjct: 317 LKARFPILMSA--PSYPLQTQVKLVVAACALHNYIRREKPDDWLFKMYEEGSFPMEESQP 490
Query: 293 ----DLIAEVDAELANQNVSHDNHEASRSDMDEFAQGGIIKNGAAHQMWSNY 340
++ ++D E Q H D +E A +++ A +MW+++
Sbjct: 491 PLEMEVPPKMDVETQTQPSVH------TFDSEEIALASQLRDSIATEMWNDF 628
>TC210984
Length = 1085
Score = 69.7 bits (169), Expect = 2e-12
Identities = 42/119 (35%), Positives = 59/119 (49%), Gaps = 6/119 (5%)
Frame = +3
Query: 230 KELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAE 289
KELFNLRHASLRN IER FG+ K RF I ++ P + K Q ++ AC LHN+L
Sbjct: 423 KELFNLRHASLRNVIERIFGIFKSRFTIFKSA--PPFLFKTQAELVLACAALHNFLRKEC 596
Query: 290 PNEDLIAEVDAELANQ-----NVSHDNHE-ASRSDMDEFAQGGIIKNGAAHQMWSNYKN 342
+++ E E ++ N ++HE ++ E I + MW N N
Sbjct: 597 RSDEFPVEPTDESSSSSSVLPNYEDNDHEPIVQTQEQEREDANIWRTNIGSDMWRNANN 773
>AW396897 similar to GP|28416671|gb| At5g41980 {Arabidopsis thaliana},
partial (24%)
Length = 430
Score = 66.6 bits (161), Expect = 2e-11
Identities = 34/70 (48%), Positives = 44/70 (62%)
Frame = +2
Query: 15 RMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGETIS 74
RM F KLCD+L+ +G LR T +EEQ+ ++I+ HN + R V FR SGETIS
Sbjct: 221 RMDKHVFYKLCDILQAKGLLRHTNRIKIEEQLGIFMFIIGHNLRTRAVQELFRYSGETIS 400
Query: 75 RHLHQVLKAI 84
RH + VL AI
Sbjct: 401 RHFNNVLNAI 430
>TC231143
Length = 941
Score = 59.7 bits (143), Expect = 2e-09
Identities = 33/101 (32%), Positives = 55/101 (53%), Gaps = 2/101 (1%)
Frame = +3
Query: 52 ILTHNAKNREVNFWFRRSGETISRHLHQVLKAILELEEKFI--VQPDGSTIPLEISSSTR 109
++ H N+ + F S E +SRHLH+VL A+L+L + + P +I + T
Sbjct: 639 LVDHVKGNKNLQERF*DSEENMSRHLHEVLIALLKLSMEXXRPLDPLFRDSHAKIENDTW 818
Query: 110 FYPYFKDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPTQNVL 150
++P+FK+ +G DGTH+ + R+ RK PTQN++
Sbjct: 819 YWPFFKNVIGTNDGTHVPCMXXHSEQARFIRRKGIPTQNIM 941
>CO984808
Length = 749
Score = 33.9 bits (76), Expect(2) = 5e-06
Identities = 33/124 (26%), Positives = 49/124 (38%)
Frame = -3
Query: 200 FMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILS 259
F S L+TPY G K FS N + FN R + ++A LK ++I+
Sbjct: 459 FPFFSWLLTPYEG-----KGFS------NVQVEFNKRVVETQMVAKKALARLKDMWKIIQ 313
Query: 260 NSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAELANQNVSHDNHEASRSDM 319
K ++I+ CCILHN ++ E D L + H + +
Sbjct: 312 GVMWKPDKHKLPRIIL-VCCILHNIVIDME---------DEVLIDMPSCHQHDSRYQDQT 163
Query: 320 DEFA 323
EFA
Sbjct: 162 SEFA 151
Score = 33.9 bits (76), Expect(2) = 5e-06
Identities = 26/99 (26%), Positives = 47/99 (47%), Gaps = 16/99 (16%)
Frame = -2
Query: 120 AIDGTHIRVKVSAKDAPR--YRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIK 177
A+D THI + + DA + R++ + + A +F ++ GW GS SD ++++
Sbjct: 748 AVDSTHIMMTLXXVDAXNSVWLDREKNCSMVLQAIVXPXXRFRDIVTGWPGSMSDEQVLR 569
Query: 178 NA----LTRE------DKLKIPEG----KYYLVDAGFML 202
++ L E K +P+G +Y + D GF L
Sbjct: 568 SSSFFKLAEEGKRLNGGKKTLPDGTLFREYIIGDTGFPL 452
>TC207824 similar to GB|AAN73294.1|25141199|BT002297 At5g12010/F14F18_180
{Arabidopsis thaliana;} , partial (38%)
Length = 847
Score = 47.4 bits (111), Expect = 1e-05
Identities = 37/128 (28%), Positives = 60/128 (45%), Gaps = 3/128 (2%)
Frame = +3
Query: 159 FTYVLAGWEGSASDSRII-KNALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHL 217
FT V GW GS D +++ K+AL + + +G + + +G+ L ++ PY
Sbjct: 66 FTDVCIGWPGSMPDDQVLEKSALFQRANGGLLKGDWIVGSSGYPLMDWVLVPY------- 224
Query: 218 KEFSARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKL--II 275
S +N + FN + ++ AF LK R+ L TE +K Q L ++
Sbjct: 225 ---SQQNLTWT-QHAFNEKIGEVQKVARDAFARLKGRWSCLQKRTE----VKLQDLPVVL 380
Query: 276 FACCILHN 283
ACC+LHN
Sbjct: 381 GACCVLHN 404
>TC228497
Length = 779
Score = 45.4 bits (106), Expect = 4e-05
Identities = 34/128 (26%), Positives = 61/128 (47%), Gaps = 3/128 (2%)
Frame = +2
Query: 159 FTYVLAGWEGSASDSRII-KNALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHL 217
F+ V GW GS SD +++ K+AL + + + + + ++G L G++ PY
Sbjct: 104 FSDVCIGWPGSLSDDQVLEKSALYQRATMGNLKDVWVVGNSGHPLMEGVLVPYTHQNLTW 283
Query: 218 KEFSARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKL--II 275
+ + FN + +++ + AF LK R+ L TE +K + L ++
Sbjct: 284 TQHA-----------FNQKVGEIQSIAKDAFARLKGRWSCLQKRTE----VKLEDLPVLL 418
Query: 276 FACCILHN 283
ACC+LHN
Sbjct: 419 GACCVLHN 442
>BE607970
Length = 485
Score = 43.9 bits (102), Expect = 1e-04
Identities = 38/145 (26%), Positives = 65/145 (44%), Gaps = 16/145 (11%)
Frame = +1
Query: 158 KFTYVLAGWEGSAS------DSRIIKNALTREDKLKIP-----EG----KYYLVDAGFML 202
+F V AGW + +S++ + ++ L P EG +Y L D+ F L
Sbjct: 61 RFLDVSAGWPSTMKPEIFLRESKLYREVEQSKELLNGPSYNLSEGCLIPQYILGDSCFPL 240
Query: 203 TSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNST 262
L+TPY V SA + FN H + + AFG L+ R+ +L+ S
Sbjct: 241 LPWLLTPYNRVNEEDSFGSA-------ERAFNCVHGNAMELVGDAFGRLRARWRLLAASR 399
Query: 263 E-PAYGIKAQKLIIFACCILHNYLM 286
+ ++ ++ ACC+LHN+++
Sbjct: 400 KWKQECVEHLPFVVVACCLLHNFVV 474
>TC223380 weakly similar to GB|AAP80175.1|32362295|BT009674 At3g55350
{Arabidopsis thaliana;} , partial (20%)
Length = 445
Score = 41.2 bits (95), Expect = 7e-04
Identities = 39/142 (27%), Positives = 66/142 (46%), Gaps = 2/142 (1%)
Frame = +3
Query: 197 DAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFE 256
D+G+ L L+ PY G KE S K FN +H + R +RA LK+ +
Sbjct: 12 DSGYRLLPYLVVPYEG-----KELSEP------KAQFNKQHLATRMVAQRALARLKEMWR 158
Query: 257 ILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAELANQNVSHDN--HEA 314
I+ + ++I+ CC+LHN ++ DL EV EL + ++ HD+ H
Sbjct: 159 IIHGMMWRPDKHRLPRIIL-VCCVLHNIVI------DLNDEVQDEL-SLSLDHDSGYHRL 314
Query: 315 SRSDMDEFAQGGIIKNGAAHQM 336
+DE +G +++ +H +
Sbjct: 315 VCGAVDE--KGVALRDRLSHYL 374
>TC212508
Length = 752
Score = 38.5 bits (88), Expect = 0.005
Identities = 33/128 (25%), Positives = 55/128 (42%), Gaps = 3/128 (2%)
Frame = +2
Query: 159 FTYVLAGWEGSASDSRII-KNALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHL 217
F V G GS R++ K+AL + +G + + ++G L ++ PY
Sbjct: 53 FNDVCIGXXGSXXXDRVLEKSALFQRASRGNLKGVWIVXNSGHPLMDWVLVPYTHANLTW 232
Query: 218 KEFSARNPPLNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKL--II 275
+ + FN + ++ + AF LK R+ L TE +K Q L ++
Sbjct: 233 TQHA-----------FNEKIEEIQGVAKEAFARLKGRWGCLQKRTE----VKLQDLPVVL 367
Query: 276 FACCILHN 283
ACC+LHN
Sbjct: 368 GACCVLHN 391
>TC219936 similar to UP|Q9LJL8 (Q9LJL8) Emb|CAB43653.1 (AT3g19120/MVI11_3),
partial (43%)
Length = 835
Score = 35.4 bits (80), Expect = 0.038
Identities = 32/108 (29%), Positives = 46/108 (41%), Gaps = 1/108 (0%)
Frame = +1
Query: 193 YYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVLK 252
Y + D F L L+TP FS + LF+ R+ + A +LK
Sbjct: 178 YVVGDWCFPLLPFLLTP----------FSPSGMGTPAQNLFDGMLMKGRSVVVEAIALLK 327
Query: 253 KRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSA-EPNEDLIAEVD 299
R++IL + G++ I ACC+LHN A EP +L E D
Sbjct: 328 GRWKILQDLNT---GVRHAPQTIVACCVLHNLCQIAREPEPELWKEPD 462
>TC231938 similar to UP|Q9LJL8 (Q9LJL8) Emb|CAB43653.1 (AT3g19120/MVI11_3),
partial (40%)
Length = 565
Score = 33.5 bits (75), Expect = 0.15
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Frame = +2
Query: 241 RNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSA-EPNEDLIAEVD 299
R+ + A +LK R++IL + G+ I ACC+LHN A EP +L E D
Sbjct: 281 RSVVVEAIALLKGRWKILQDLNT---GVHHAPQTIVACCVLHNLCQIAREPEPELWKEPD 451
>TC222807 similar to UP|Q9FK34 (Q9FK34) Gb|AAD25781.1, partial (13%)
Length = 399
Score = 28.9 bits (63), Expect = 3.6
Identities = 30/114 (26%), Positives = 46/114 (40%), Gaps = 3/114 (2%)
Frame = -2
Query: 223 RNPPLNYKELFNLRHASLRN---AIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACC 279
R PPLN LFN + + N I+ LS T P + I ++
Sbjct: 377 RTPPLNLLNLFNTQPLNTENNQTLIQSVTTDAAINL*TLSLITRPNHAIDDFSEVL---- 210
Query: 280 ILHNYLMSAEPNEDLIAEVDAELANQNVSHDNHEASRSDMDEFAQGGIIKNGAA 333
N L + + +E ++AE + EA+R D+ E +GGI+ G A
Sbjct: 209 ---NGLGTHQIDEQVVAEARGAVV---------EAARVDLAEALEGGIVNCGGA 84
>TC228846 similar to UP|Q9LMV4 (Q9LMV4) F5M15.23, partial (6%)
Length = 645
Score = 28.9 bits (63), Expect = 3.6
Identities = 29/119 (24%), Positives = 48/119 (39%), Gaps = 23/119 (19%)
Frame = +2
Query: 13 IIRMGPQTFLKLCDMLEREGGLRPTRW--SSVEEQVAKSLYIL---------THNAKNRE 61
I ++ P TF+ +CD ++ G RW +S+ V +S ++L N + R
Sbjct: 11 ITQVNPSTFIAICDQRGKKAGRFCCRWVSNSIGRVVDRSAWLLLQLLLDRAPPRNIRGRY 190
Query: 62 VNFWFRRS-GETISRH-----------LHQVLKAILELEEKFIVQPDGSTIPLEISSST 108
W RS G T S + +++ + K P GST L ++ST
Sbjct: 191 *GLWRTRSNGRTASSNSSP*RVSFY*ACDL*VRSTKSMASKKTSTPSGSTTVLSPTAST 367
>TC210238 similar to UP|Q9SW27 (Q9SW27) Geranylgeranylated protein ATGP4
(At4g24990), partial (97%)
Length = 690
Score = 28.5 bits (62), Expect = 4.7
Identities = 13/50 (26%), Positives = 24/50 (48%)
Frame = -3
Query: 248 FGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAE 297
FG L KR+ L+N + + CC+ +N+L+S ++ + E
Sbjct: 439 FGKLAKRYSTLANCLVVFQNFASTYQLYIICCLWYNFLVSGPVCKNAVLE 290
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,443,187
Number of Sequences: 63676
Number of extensions: 180321
Number of successful extensions: 872
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 862
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 862
length of query: 346
length of database: 12,639,632
effective HSP length: 98
effective length of query: 248
effective length of database: 6,399,384
effective search space: 1587047232
effective search space used: 1587047232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146752.6


