

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.2 + phase: 0
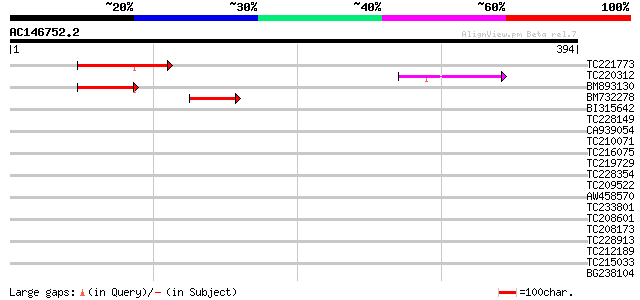
(394 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221773 similar to GB|AAP37835.1|30725626|BT008476 At1g07010 {A... 75 7e-14
TC220312 similar to GB|AAP37835.1|30725626|BT008476 At1g07010 {A... 53 2e-07
BM893130 similar to GP|19310495|gb| At1g07010/F10K1_19 {Arabidop... 46 3e-05
BM732278 similar to GP|19310495|gb| At1g07010/F10K1_19 {Arabidop... 42 4e-04
BI315642 37 0.012
TC228149 similar to UP|O02402 (O02402) Insoluble protein, partia... 36 0.026
CA939054 similar to GP|11994773|db phosphoprotein phosphatase {A... 35 0.077
TC210071 homologue to UP|BSL1_ARATH (Q8L7U5) Serine/threonine pr... 34 0.10
TC216075 UP|Q8LSN3 (Q8LSN3) Serine/threonine protein phosphatase... 33 0.17
TC219729 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 32 0.50
TC228354 homologue to UP|BSL2_ARATH (Q9SJF0) Serine/threonine pr... 32 0.50
TC209522 similar to UP|O22600 (O22600) Glycine-rich protein, par... 32 0.50
AW458570 similar to GP|11994255|dbj contains similarity to termi... 32 0.50
TC233801 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 32 0.50
TC208601 32 0.50
TC208173 similar to GB|AAS92332.1|46402460|BT012416 At3g08630 {A... 32 0.65
TC228913 weakly similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partia... 32 0.65
TC212189 weakly similar to UP|Q42421 (Q42421) Chitinase precurso... 32 0.65
TC215033 similar to GB|AAL05900.1|15777879|AY055100 AT3g15640/MS... 32 0.65
BG238104 similar to GP|11994336|dbj cytochrome c oxidase subunit... 32 0.65
>TC221773 similar to GB|AAP37835.1|30725626|BT008476 At1g07010 {Arabidopsis
thaliana;} , partial (24%)
Length = 449
Score = 74.7 bits (182), Expect = 7e-14
Identities = 37/69 (53%), Positives = 52/69 (74%), Gaps = 3/69 (4%)
Frame = +3
Query: 48 PTRLPSPSR-LIAIGDLHGDLKKSKEALSIAGLIDSSGN--YTGGSATVVQIGDVLDRGG 104
PT + +P R ++A+GDLHGDLK+++ AL +AG++ S G +TGG +VQ+GD+LDRG
Sbjct: 237 PTFVSAPGRRILAVGDLHGDLKQARSALEMAGVLSSDGQDLWTGGETVLVQLGDILDRGE 416
Query: 105 DEIKILYLL 113
DEI IL LL
Sbjct: 417 DEIAILSLL 443
>TC220312 similar to GB|AAP37835.1|30725626|BT008476 At1g07010 {Arabidopsis
thaliana;} , partial (29%)
Length = 641
Score = 53.1 bits (126), Expect = 2e-07
Identities = 33/84 (39%), Positives = 43/84 (50%), Gaps = 9/84 (10%)
Frame = +2
Query: 271 RGRNALVWLRKFSDGNCD---------CSSLEHVLSTIPGVKRMIMGHTIQKEGINGVCE 321
RG +++VW R +S D CS L+ L + G K M++GHT Q G+N
Sbjct: 20 RGYDSVVWNRLYSRDTLDLVDYQDKQVCSILDETLQAV-GAKAMVVGHTPQTIGVNCKYN 196
Query: 322 NKAIRIDVGMSKGCGGGLPEVLEI 345
R+DVGMS G PEVLEI
Sbjct: 197 CSIWRVDVGMSSGVLNSRPEVLEI 268
>BM893130 similar to GP|19310495|gb| At1g07010/F10K1_19 {Arabidopsis
thaliana}, partial (13%)
Length = 421
Score = 46.2 bits (108), Expect = 3e-05
Identities = 22/45 (48%), Positives = 34/45 (74%), Gaps = 3/45 (6%)
Frame = +1
Query: 48 PTRLPSPSR-LIAIGDLHGDLKKSKEALSIAGLIDSSGN--YTGG 89
PT + +P R ++A+GDLHGDLK+++ AL +AG++ S G +TGG
Sbjct: 286 PTFVSAPGRRILAVGDLHGDLKQARSALEMAGVLSSDGRDLWTGG 420
>BM732278 similar to GP|19310495|gb| At1g07010/F10K1_19 {Arabidopsis
thaliana}, partial (7%)
Length = 422
Score = 42.4 bits (98), Expect = 4e-04
Identities = 18/35 (51%), Positives = 24/35 (68%)
Frame = +1
Query: 126 NFITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEW 160
+FI +NGNHE MN EGDFR+ G +E +LE+
Sbjct: 22 HFI*VNGNHETMNVEGDFRYVDSGGFDECNDFLEY 126
>BI315642
Length = 141
Score = 37.4 bits (85), Expect = 0.012
Identities = 23/39 (58%), Positives = 26/39 (65%)
Frame = +1
Query: 356 NPLYNQMNKENVDIGKVEEGFGLLLNNQDGRPRQVEVKA 394
NPLY NK NVD+GK E+G G + G PRQVEVKA
Sbjct: 4 NPLYQ--NKGNVDVGK-EQGLG-----EHGGPRQVEVKA 96
>TC228149 similar to UP|O02402 (O02402) Insoluble protein, partial (4%)
Length = 1008
Score = 36.2 bits (82), Expect = 0.026
Identities = 40/142 (28%), Positives = 58/142 (40%), Gaps = 16/142 (11%)
Frame = +1
Query: 35 LFLPPPPSSPPPIPTRLP------SPSRLIAIGDLHGD---LKKSKEALSIAGLIDSSGN 85
L LPPPP PPP+P LP RLI L AL +GL S G
Sbjct: 508 LLLPPPPPPPPPLPPPLPPFDASAEVLRLIRFRKSAAALLFLSAPPSALLFSGLTVSEGL 687
Query: 86 YTGGSATVVQIGDVLDRGGDEIKIL------YLLEKLKRQAAIHGGNFITMNGNHEIMNA 139
+ A V+I D G + +L +LL +LK A + G + + + +
Sbjct: 688 FHEDPALGVEIED-----GFLLLLLLLLLLRFLLLRLKLLAPVTGFSLALGSSSSSVSEE 852
Query: 140 EGDF-RFATKNGVEEFKVWLEW 160
EG FA+++G+ + W
Sbjct: 853 EGFMGGFASEDGLSFLSLKTGW 918
>CA939054 similar to GP|11994773|db phosphoprotein phosphatase {Arabidopsis
thaliana}, partial (39%)
Length = 407
Score = 34.7 bits (78), Expect = 0.077
Identities = 25/84 (29%), Positives = 39/84 (45%)
Frame = +3
Query: 52 PSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILY 111
P + + GD+HG E +AG + ++ NY + IGD +DRG + ++
Sbjct: 135 PVSAPVTVCGDIHGQFYDLLELFRVAGEMPTT-NY-------IFIGDFVDRGYNSVETFE 290
Query: 112 LLEKLKRQAAIHGGNFITMNGNHE 135
LL LK + H + GNHE
Sbjct: 291 LLMCLKVKYPAH---ITLLRGNHE 353
>TC210071 homologue to UP|BSL1_ARATH (Q8L7U5) Serine/threonine protein
phosphatase BSL1 (BSU1-like protein 1) , partial (27%)
Length = 765
Score = 34.3 bits (77), Expect = 0.10
Identities = 27/98 (27%), Positives = 42/98 (42%)
Frame = +2
Query: 48 PTRLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEI 107
PT L + + GDLHG G ++G+ T + +GD +DRG +
Sbjct: 122 PTVLQLKAPVKVFGDLHGQFGDLMRLFDEYGFPSTAGDIT--YIDYLFLGDYVDRGQHSL 295
Query: 108 KILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRF 145
+ + LL LK + + N + GNHE + F F
Sbjct: 296 ETITLLLALKIE---YPENVHLIRGNHEAADINALFGF 400
>TC216075 UP|Q8LSN3 (Q8LSN3) Serine/threonine protein phosphatase 2A,
complete
Length = 1201
Score = 33.5 bits (75), Expect = 0.17
Identities = 23/84 (27%), Positives = 38/84 (44%)
Frame = +1
Query: 52 PSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILY 111
P S + GD+HG + G + + NY + +GD +DRG + +++
Sbjct: 223 PVNSPVTVCGDIHGQFHDLMKLFQTGGHVPET-NY-------IFMGDFVDRGYNSLEVFT 378
Query: 112 LLEKLKRQAAIHGGNFITMNGNHE 135
+L LK A + N + GNHE
Sbjct: 379 ILLLLK---ARYPANITLLRGNHE 441
>TC219729 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (7%)
Length = 785
Score = 32.0 bits (71), Expect = 0.50
Identities = 12/17 (70%), Positives = 12/17 (70%)
Frame = +3
Query: 38 PPPPSSPPPIPTRLPSP 54
PPPPS PPPIP P P
Sbjct: 198 PPPPSPPPPIPPSPPPP 248
>TC228354 homologue to UP|BSL2_ARATH (Q9SJF0) Serine/threonine protein
phosphatase BSL2 (BSU1-like protein 2) , partial (42%)
Length = 1946
Score = 32.0 bits (71), Expect = 0.50
Identities = 27/103 (26%), Positives = 44/103 (42%)
Frame = +2
Query: 43 SPPPIPTRLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDR 102
S P +L +P ++ GDLHG G ++G+ + +GD +DR
Sbjct: 437 SSEPSVLQLRAPIKIF--GDLHGQFGDLMRLFDEYGAPSTAGDIA--YIDYLFLGDYVDR 604
Query: 103 GGDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRF 145
G ++ + LL LK + + N + GNHE + F F
Sbjct: 605 GQHSLETITLLLALKVE---YPNNVHLIRGNHEAADINALFGF 724
>TC209522 similar to UP|O22600 (O22600) Glycine-rich protein, partial (47%)
Length = 717
Score = 32.0 bits (71), Expect = 0.50
Identities = 11/18 (61%), Positives = 14/18 (77%)
Frame = -2
Query: 39 PPPSSPPPIPTRLPSPSR 56
PPPS PPPIP +P+P +
Sbjct: 263 PPPSPPPPIPPPIPTPPK 210
>AW458570 similar to GP|11994255|dbj contains similarity to terminal
ear1~gene_id:MJL14.2 {Arabidopsis thaliana}, partial
(11%)
Length = 425
Score = 32.0 bits (71), Expect = 0.50
Identities = 24/64 (37%), Positives = 27/64 (41%), Gaps = 10/64 (15%)
Frame = +3
Query: 27 VDFSVSGG----LFLPPPPSS------PPPIPTRLPSPSRLIAIGDLHGDLKKSKEALSI 76
++FS GG F PPS PPP P PSP R A LH KKS +
Sbjct: 234 IEFSRPGGHTRKFFHHSPPSETTPFNVPPPPPPFPPSPRRRFAAPRLHSSQKKSPGSHKS 413
Query: 77 AGLI 80
G I
Sbjct: 414 TGSI 425
>TC233801 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (5%)
Length = 394
Score = 32.0 bits (71), Expect = 0.50
Identities = 12/17 (70%), Positives = 12/17 (70%)
Frame = +2
Query: 38 PPPPSSPPPIPTRLPSP 54
PPPPS PPPIP P P
Sbjct: 287 PPPPSPPPPIPPSPPPP 337
Score = 27.7 bits (60), Expect = 9.4
Identities = 10/17 (58%), Positives = 11/17 (63%)
Frame = +2
Query: 38 PPPPSSPPPIPTRLPSP 54
P PPS PPP+P P P
Sbjct: 311 PIPPSPPPPLPASPPPP 361
>TC208601
Length = 597
Score = 32.0 bits (71), Expect = 0.50
Identities = 24/86 (27%), Positives = 42/86 (47%), Gaps = 1/86 (1%)
Frame = +1
Query: 52 PSPSRLIAIGDLHGDLKKSKEA-LSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKIL 110
P +I +GD+HG + K + ++ G +D S T AT++ +GD DRG +++
Sbjct: 97 PKTRAVICVGDIHGFITKLQSLWKNLEGSLDRSEFET---ATLIFLGDYCDRGPATRQVI 267
Query: 111 YLLEKLKRQAAIHGGNFITMNGNHEI 136
L L + F+ GNH++
Sbjct: 268 DFLISLPSRYPRQKHVFLC--GNHDL 339
>TC208173 similar to GB|AAS92332.1|46402460|BT012416 At3g08630 {Arabidopsis
thaliana;} , partial (12%)
Length = 449
Score = 31.6 bits (70), Expect = 0.65
Identities = 15/32 (46%), Positives = 18/32 (55%)
Frame = -2
Query: 24 DTFVDFSVSGGLFLPPPPSSPPPIPTRLPSPS 55
D +DF S PPPP PPP+P P+PS
Sbjct: 328 DESLDFPSSSPQLSPPPP--PPPLPPPRPTPS 239
>TC228913 weakly similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partial (19%)
Length = 692
Score = 31.6 bits (70), Expect = 0.65
Identities = 11/20 (55%), Positives = 14/20 (70%)
Frame = +3
Query: 38 PPPPSSPPPIPTRLPSPSRL 57
PPPP PPP P+ P P+R+
Sbjct: 33 PPPPDEPPPQPSLRPHPARV 92
>TC212189 weakly similar to UP|Q42421 (Q42421) Chitinase precursor , partial
(9%)
Length = 774
Score = 31.6 bits (70), Expect = 0.65
Identities = 15/38 (39%), Positives = 18/38 (46%)
Frame = +1
Query: 38 PPPPSSPPPIPTRLPSPSRLIAIGDLHGDLKKSKEALS 75
P PPS+PPP P R P + A G L S + S
Sbjct: 439 PSPPSTPPPPPPRASPPPSVAATGSAPRSLSSSSTSSS 552
>TC215033 similar to GB|AAL05900.1|15777879|AY055100 AT3g15640/MSJ11_4
{Arabidopsis thaliana;} , partial (60%)
Length = 923
Score = 31.6 bits (70), Expect = 0.65
Identities = 11/17 (64%), Positives = 12/17 (69%)
Frame = +3
Query: 38 PPPPSSPPPIPTRLPSP 54
PPPP PPP+P LP P
Sbjct: 222 PPPPPPPPPLPDLLPPP 272
>BG238104 similar to GP|11994336|dbj cytochrome c oxidase subunit Vb
precursor-like protein {Arabidopsis thaliana}, partial
(40%)
Length = 476
Score = 31.6 bits (70), Expect = 0.65
Identities = 11/17 (64%), Positives = 12/17 (69%)
Frame = +2
Query: 38 PPPPSSPPPIPTRLPSP 54
PPPP PPP+P LP P
Sbjct: 107 PPPPPPPPPLPDLLPPP 157
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.141 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,306,215
Number of Sequences: 63676
Number of extensions: 317967
Number of successful extensions: 9198
Number of sequences better than 10.0: 223
Number of HSP's better than 10.0 without gapping: 4616
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7766
length of query: 394
length of database: 12,639,632
effective HSP length: 99
effective length of query: 295
effective length of database: 6,335,708
effective search space: 1869033860
effective search space used: 1869033860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146752.2


