

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146751.5 + phase: 0 /pseudo
(455 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
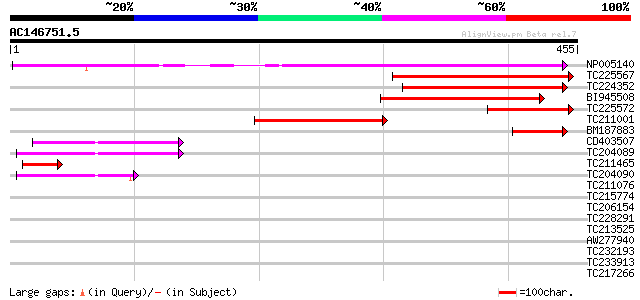
Score E
Sequences producing significant alignments: (bits) Value
NP005140 heat shock protein 232 3e-61
TC225567 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp pro... 174 1e-43
TC224352 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein... 128 6e-30
BI945508 homologue to PIR|T51523|T51 clpB heat shock protein-lik... 114 7e-26
TC225572 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp pro... 107 9e-24
TC211001 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein... 104 9e-23
BM187883 homologue to GP|9651530|gb| ClpB {Phaseolus lunatus}, p... 54 2e-07
CD403507 52 7e-07
TC204089 47 2e-05
TC211465 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp prote... 44 2e-04
TC204090 42 4e-04
TC211076 homologue to UP|Q948P2 (Q948P2) Replication factor C 40... 34 0.16
TC215774 similar to UP|Q94ES0 (Q94ES0) AAA-metalloprotease FtsH,... 33 0.20
TC206154 homologue to UP|FTSH_MEDSA (Q9BAE0) Cell division prote... 31 1.3
TC228291 homologue to UP|Q9FXT8 (Q9FXT8) 26S proteasome regulato... 30 2.9
TC213525 similar to UP|Q8JKS7 (Q8JKS7) Orf34, partial (7%) 29 3.8
AW277940 similar to GP|7573596|dbj| protein kinase 1 {Populus ni... 29 3.8
TC232193 weakly similar to UP|Q94AF3 (Q94AF3) AT4g28740/F16A16_1... 29 5.0
TC233913 weakly similar to UP|O22429 (O22429) Calcium binding pr... 28 6.6
TC217266 similar to GB|AAL99238.1|22651517|AY082345 short-chain ... 28 6.6
>NP005140 heat shock protein
Length = 2736
Score = 232 bits (591), Expect = 3e-61
Identities = 148/449 (32%), Positives = 237/449 (51%), Gaps = 4/449 (0%)
Frame = +1
Query: 3 ENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR-- 60
E T + + +A + A GH ++ H+ ++ D NG+ + S G +R
Sbjct: 10 EKFTHKTNEALASAHELAMSSGHAQLTPIHLAHALISDPNGIFVLAINSAGGGEESARAV 189
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
E+V+ + C ++ + + + + KS G + L+LG+L+ S
Sbjct: 190 ERVLNQALKKLPCQSPPPDEVPASTNLVRAIRRAQAAQKSRGDTRLAVDQLILGILEDSQ 369
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
+ L++ G V K+ +V +++ G+ + V + S +N
Sbjct: 370 --IGDLLKEAGVAVAKVESEV-------------------DKLRGKEGKKVESASGDTNF 486
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
AL+ +G +L + Q GKL P +GR+E++ RV++I+
Sbjct: 487 ------------------------QALKTYGRDLVE--QAGKLDPVIGRDEEIRRVVRIL 588
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP LVGEPGVGKT++++GLAQRI+ G VP L +++ALD+ + +G
Sbjct: 589 SRRTKNNPVLVGEPGVGKTAVVEGLAQRIVRGDVPSNLADVRLIALDMGALVAGAKYRGE 768
Query: 301 SEERIRCLIKEIELC-GNVILFVKEVHHIFDAA-TSGARSFAYILKHALERGVIQCIFAT 358
EER++ ++KE+E G VILF+ E+H + A T G+ A + K L RG ++CI AT
Sbjct: 769 FEERLKAVLKEVEEAEGKVILFIDEIHLVLGAGRTEGSMDAANLFKPMLARGQLRCIGAT 948
Query: 359 TVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAA 418
T+ E+R ++E D +R FQ V V EPSV +TI IL+GL+ YE H+ + D ALV AA
Sbjct: 949 TLEEYRKYVEKDAAFERRFQQVFVAEPSVVDTISILRGLKERYEGHHGVRIQDRALVMAA 1128
Query: 419 NLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS +Y++ R LPDKAIDL+DEA ++V++
Sbjct: 1129QLSNRYITGRHLPDKAIDLVDEACANVRV 1215
>TC225567 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (61%)
Length = 2077
Score = 174 bits (440), Expect = 1e-43
Identities = 92/146 (63%), Positives = 113/146 (77%), Gaps = 1/146 (0%)
Frame = +3
Query: 308 LIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATTVNEHRMH 366
L++EI+ +ILF+ EVH + A A GA A ILK AL RG +QCI ATT++E+R H
Sbjct: 12 LMEEIKQSDEIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKH 191
Query: 367 MENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANLSQQYVS 426
+E D L+R FQPVKV EP+V+ETI+ILKGLR YE H+KL YTDEALVAAA LS QY+S
Sbjct: 192 IEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDEALVAAAQLSYQYIS 371
Query: 427 ERFLPDKAIDLIDEAGSHVQLCHAKV 452
+RFLPDKAIDLIDEAGS V+L HA++
Sbjct: 372 DRFLPDKAIDLIDEAGSRVRLQHAQL 449
>TC224352 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like,
partial (17%)
Length = 505
Score = 128 bits (321), Expect = 6e-30
Identities = 67/133 (50%), Positives = 94/133 (70%), Gaps = 1/133 (0%)
Frame = +1
Query: 316 GNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATTVNEHRMHMENDTTLK 374
G ILF+ E+H + A A++GA +LK L RG ++CI ATT++E+R ++E D L+
Sbjct: 1 GQTILFIDEIHTVVGAGASNGAMDAGNLLKPMLGRGELRCIGATTLDEYRKYIEKDPALE 180
Query: 375 RIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANLSQQYVSERFLPDKA 434
R FQ V V +PSVE+TI IL+GLR YE H+ + +D ALV AA LS +Y+S RFLPDKA
Sbjct: 181 RRFQQVYVDQPSVEDTISILRGLRERYELHHGVRISDSALVDAAILSDRYISGRFLPDKA 360
Query: 435 IDLIDEAGSHVQL 447
IDL+DEA + +++
Sbjct: 361 IDLVDEAAAKLKM 399
>BI945508 homologue to PIR|T51523|T51 clpB heat shock protein-like -
Arabidopsis thaliana, partial (14%)
Length = 432
Score = 114 bits (286), Expect = 7e-26
Identities = 62/134 (46%), Positives = 91/134 (67%), Gaps = 2/134 (1%)
Frame = +1
Query: 298 QGSSEERIRCLIKEI-ELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCI 355
+G E+R++ ++KE+ E G ILF+ E+H + A AT+GA +LK L RG ++CI
Sbjct: 31 RGEFEDRLKAVLKEVTESDGQTILFIDEIHTVVGAGATNGAMDAGNLLKPMLGRGELRCI 210
Query: 356 FATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALV 415
ATT++E+R ++E D L+R FQ V V +P+VE+TI IL+GLR YE H+ + +D ALV
Sbjct: 211 GATTLDEYRKYIEKDPALERRFQQVYVDQPTVEDTISILRGLRERYELHHGVRISDSALV 390
Query: 416 AAANLSQQYVSERF 429
AA LS +Y+S RF
Sbjct: 391 EAAILSDRYISGRF 432
>TC225572 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (18%)
Length = 511
Score = 107 bits (268), Expect = 9e-24
Identities = 52/69 (75%), Positives = 61/69 (88%)
Frame = +3
Query: 384 EPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANLSQQYVSERFLPDKAIDLIDEAGS 443
EP+V ETI+ILKGLR YE H+KLHYTD+ALVAAA LS QY+S+RFLPDKAIDLIDEAGS
Sbjct: 12 EPTVNETIQILKGLRERYEIHHKLHYTDDALVAAAQLSHQYISDRFLPDKAIDLIDEAGS 191
Query: 444 HVQLCHAKV 452
V+L HA++
Sbjct: 192 RVRLQHAQL 218
>TC211001 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like,
partial (16%)
Length = 456
Score = 104 bits (259), Expect = 9e-23
Identities = 48/107 (44%), Positives = 74/107 (68%)
Frame = +2
Query: 197 DDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICRRMKNNPCLVGEPGV 256
D + + ALE +G +LT +A+ GKL P +GR++++ R IQI+ RR KNNP L+GEPGV
Sbjct: 134 DQDPEGKYEALEKYGKDLTAMAKAGKLDPVIGRDDEIRRCIQILSRRTKNNPVLIGEPGV 313
Query: 257 GKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEE 303
GKT+I +GLAQRI+ G VP+ L +++++LD+ + +G E+
Sbjct: 314 GKTAISEGLAQRIVQGDVPQALMNRRLISLDMGALIAGAKYRGEFED 454
>BM187883 homologue to GP|9651530|gb| ClpB {Phaseolus lunatus}, partial (14%)
Length = 427
Score = 53.5 bits (127), Expect = 2e-07
Identities = 23/44 (52%), Positives = 36/44 (81%)
Frame = +3
Query: 404 HYKLHYTDEALVAAANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
H+ + +D ALV+AA L+ +Y++ERFLPDKAIDL+DEA + +++
Sbjct: 3 HHGVKISDSALVSAAVLADRYITERFLPDKAIDLVDEAAAKLKM 134
>CD403507
Length = 633
Score = 51.6 bits (122), Expect = 7e-07
Identities = 39/123 (31%), Positives = 63/123 (50%), Gaps = 2/123 (1%)
Frame = -2
Query: 19 EARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQVMKLVGRGGGCSGFSC 78
EAR + + TE IL+GIL + AAK L++ GI +RE+ ++L+G+ FS
Sbjct: 620 EARKLKYPNTGTEAILMGILVEGTSNAAKFLRANGITLLKAREETVELLGK-SDLFFFSP 444
Query: 79 KDIRFTFDAKNVLDFSLKH-AKSLGY-DNVDTMHLLLGLLQGSNGTVTQLIQNQGADVNK 136
+ T A+ LD++++ KS G ++ HLLLG+ Q++ G D K
Sbjct: 443 EHPPLTEPAQKALDWAIEEKLKSAGEGGEINVTHLLLGIWSQEESAGQQILATLGFDDGK 264
Query: 137 IRE 139
+E
Sbjct: 263 AQE 255
Score = 29.3 bits (64), Expect = 3.8
Identities = 34/127 (26%), Positives = 53/127 (40%), Gaps = 2/127 (1%)
Frame = -2
Query: 5 LTECAKKVI-MAAQKEARFVGHK-YISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQ 62
LTE A+K + A +++ + G I+ H+LLGI +IL + G D ++E
Sbjct: 431 LTEPAQKALDWAIEEKLKSAGEGGEINVTHLLLGIWSQEESAGQQILATLGFDDGKAQEL 252
Query: 63 VMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGT 122
K+ G D+ F F + S + VD L LG+ S G
Sbjct: 251 SKKIDG-----------DV*FKFQKGQA*EISCRLM-------VD*YPLTLGVASSS*GK 126
Query: 123 VTQLIQN 129
+T L Q+
Sbjct: 125 LTILPQD 105
>TC204089
Length = 1141
Score = 46.6 bits (109), Expect = 2e-05
Identities = 38/135 (28%), Positives = 64/135 (47%), Gaps = 1/135 (0%)
Frame = +2
Query: 6 TECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQVMK 65
+E A K + EAR + + TE +L+GIL + AAK ++ GI RE+ +
Sbjct: 368 SERAIKSYAMGELEARKLKYPNTGTEALLMGILVEGTSKAAKFSRANGITLLKVREETVG 547
Query: 66 LVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKH-AKSLGYDNVDTMHLLLGLLQGSNGTVT 124
L+G+ FS + T A+ LD++++ KS ++ HLLLG+
Sbjct: 548 LLGK-SDLFFFSPEHPPLTEPAQKALDWAIEEKLKSGEGGEINATHLLLGIWSQKESAGQ 724
Query: 125 QLIQNQGADVNKIRE 139
Q++ G + K +E
Sbjct: 725 QILATLGFNDEKAKE 769
>TC211465 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (15%)
Length = 593
Score = 43.5 bits (101), Expect = 2e-04
Identities = 18/32 (56%), Positives = 26/32 (81%)
Frame = +1
Query: 11 KVIMAAQKEARFVGHKYISTEHILLGILGDSN 42
K+IM AQ+EAR +GH ++ TE ILLG++G+ N
Sbjct: 478 KLIMLAQEEARRLGHNFVGTEQILLGLIGERN 573
>TC204090
Length = 855
Score = 42.4 bits (98), Expect = 4e-04
Identities = 33/100 (33%), Positives = 52/100 (52%), Gaps = 2/100 (2%)
Frame = +3
Query: 6 TECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQVMK 65
+E A K + EAR + + TE IL+GIL + AAK L++ GI +RE+ ++
Sbjct: 243 SERAIKSYAMGELEARKLKYPNTGTEAILMGILVEGTSNAAKFLRANGITLLKAREETVE 422
Query: 66 LVGRGGGCSGFSCKDIRFTFDAKNVLDFSL--KHAKSLGY 103
L+G+ FS + T A+ LD+++ K LGY
Sbjct: 423 LLGK-SDLFFFSPEHPPLTEPAQKALDWAIEEKLKSDLGY 539
>TC211076 homologue to UP|Q948P2 (Q948P2) Replication factor C 40kDa subunit,
partial (40%)
Length = 504
Score = 33.9 bits (76), Expect = 0.16
Identities = 23/53 (43%), Positives = 30/53 (56%), Gaps = 2/53 (3%)
Frame = +3
Query: 222 KLHPFVGREEQVERVIQIICRRMKNNPCLV--GEPGVGKTSIIQGLAQRILSG 272
K+ VG E+ V R +Q+I R N P L+ G PG GKT+ I LA +L G
Sbjct: 138 KVADIVGNEDAVSR-LQVIARD-GNMPNLILSGPPGTGKTTSILALAHELLGG 290
>TC215774 similar to UP|Q94ES0 (Q94ES0) AAA-metalloprotease FtsH, partial
(79%)
Length = 2084
Score = 33.5 bits (75), Expect = 0.20
Identities = 23/79 (29%), Positives = 36/79 (45%)
Frame = +3
Query: 250 LVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRCLI 309
LVG PG GKT + + A G +++ +DF+ + G S R+R L
Sbjct: 612 LVGPPGTGKTLLAKATAGE----------SGVPFLSISGSDFMEMFVGVGPS--RVRNLF 755
Query: 310 KEIELCGNVILFVKEVHHI 328
+E C I+F+ E+ I
Sbjct: 756 QEARQCSPSIVFIDEIDAI 812
>TC206154 homologue to UP|FTSH_MEDSA (Q9BAE0) Cell division protein ftsH
homolog, chloroplast precursor , partial (88%)
Length = 2059
Score = 30.8 bits (68), Expect = 1.3
Identities = 24/87 (27%), Positives = 42/87 (47%)
Frame = +2
Query: 250 LVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRCLI 309
LVG PG GKT L R ++G G + ++F+ + G+S R+R L
Sbjct: 635 LVGPPGTGKT-----LLARAVAGEA-----GVPFFSCAASEFVELFVGVGAS--RVRDLF 778
Query: 310 KEIELCGNVILFVKEVHHIFDAATSGA 336
++ + I+F+ E+ +++A GA
Sbjct: 779 EKAKGKAPCIVFIDEIDAVWEAERGGA 859
>TC228291 homologue to UP|Q9FXT8 (Q9FXT8) 26S proteasome regulatory particle
triple-A ATPase subunit4, partial (62%)
Length = 1079
Score = 29.6 bits (65), Expect = 2.9
Identities = 46/182 (25%), Positives = 76/182 (41%), Gaps = 13/182 (7%)
Frame = +3
Query: 250 LVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRCLI 309
L G PG GKT + + +A I + + KVV+ + D Y+ G S IR +
Sbjct: 93 LYGPPGTGKTLLARAIASNIEANFL-------KVVSSAIID-KYI----GESARLIREMF 236
Query: 310 KEIELCGNVILFVKEVHHI----FDAATSGARSFAYILKHALER-------GVIQCIFAT 358
I+F+ E+ I F TS R L L + G ++ I AT
Sbjct: 237 GYARDHQPCIIFMDEIDAIGGRRFSEGTSADREIQRTLMELLNQLDGFDQLGKVKMIMAT 416
Query: 359 TVNEHRMHMENDTTLK--RIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVA 416
+R + + L+ R+ + +++ P+ + +EILK H ++ Y EA+V
Sbjct: 417 ----NRPDVLDPALLRPGRLDRKIEIPLPNEQSRMEILKIHAAGIAKHGEIDY--EAVVK 578
Query: 417 AA 418
A
Sbjct: 579 LA 584
>TC213525 similar to UP|Q8JKS7 (Q8JKS7) Orf34, partial (7%)
Length = 620
Score = 29.3 bits (64), Expect = 3.8
Identities = 21/71 (29%), Positives = 36/71 (50%), Gaps = 3/71 (4%)
Frame = +3
Query: 292 LYVISNQGSSEERIRCLIKEIELCGN---VILFVKEVHHIFDAATSGARSFAYILKHALE 348
L+ + QGS+ I CL+ ++ G+ F+ EV +++ S A IL+H+
Sbjct: 186 LFEVIFQGSN---ICCLMWKMYFIGSSCDTFFFL*EVRYLYLLLNSYCHLIA*ILRHSWT 356
Query: 349 RGVIQCIFATT 359
V+ C+F TT
Sbjct: 357 LLVVDCVFNTT 389
>AW277940 similar to GP|7573596|dbj| protein kinase 1 {Populus nigra},
partial (12%)
Length = 415
Score = 29.3 bits (64), Expect = 3.8
Identities = 21/72 (29%), Positives = 36/72 (49%), Gaps = 2/72 (2%)
Frame = +3
Query: 189 NNNLGARKDDNANK-QKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICRRMK-N 246
N+NL +K+DN K + +L++ NL +++ EGK++ G Q ++
Sbjct: 198 NSNLNGKKEDNNPKPDQLSLDVKYLNLKEVSNEGKVN---GYRAQTFTFAELAAATGNFR 368
Query: 247 NPCLVGEPGVGK 258
C +GE G GK
Sbjct: 369 LDCFLGEGGFGK 404
>TC232193 weakly similar to UP|Q94AF3 (Q94AF3) AT4g28740/F16A16_150, partial
(48%)
Length = 760
Score = 28.9 bits (63), Expect = 5.0
Identities = 39/146 (26%), Positives = 58/146 (39%), Gaps = 23/146 (15%)
Frame = +2
Query: 256 VGKTSIIQGLAQRILSGSVPEKLKGKKV--VALDVADFLYVISNQGSSEERIRCLIKEIE 313
+ T +I LA + VPE LKG + A+ + FLYV N+ + + R + E
Sbjct: 320 IATTQLIGALANSSRASEVPEILKGLGIDLAAVSLFAFLYVRENKAKNAQMAR--LSREE 493
Query: 314 LCGNVILFVKE-----VHHIFDAA----TSGARSFAY--------ILKHALERGVIQCIF 356
N+ L V E V+ + A +G SF L+RGV+ F
Sbjct: 494 SLSNLKLRVDEKKIIPVNSLRGIARLVICAGPASFVTESFKRSEPFTDSLLDRGVLVVPF 673
Query: 357 ATTVNEHRMHMEND----TTLKRIFQ 378
T N + E T KR++Q
Sbjct: 674 VTDGNSPALEFEETEELATRRKRLWQ 751
>TC233913 weakly similar to UP|O22429 (O22429) Calcium binding protein,
partial (14%)
Length = 511
Score = 28.5 bits (62), Expect = 6.6
Identities = 20/61 (32%), Positives = 30/61 (48%), Gaps = 5/61 (8%)
Frame = +3
Query: 89 NVLDFSLKHAKSLGYDN-----VDTMHLLLGLLQGSNGTVTQLIQNQGADVNKIREQVIR 143
+V+D S HA+ Y V T+H LL L + + Q +NQG D +E+ I+
Sbjct: 174 DVID*SYTHAQQSSYSQSPLIYVATLHYLLHLSS*EDQILPQ*TKNQGHDECMPQEETIQ 353
Query: 144 Q 144
Q
Sbjct: 354 Q 356
>TC217266 similar to GB|AAL99238.1|22651517|AY082345 short-chain
dehydrogenase/reductase {Arabidopsis thaliana;} ,
partial (93%)
Length = 1133
Score = 28.5 bits (62), Expect = 6.6
Identities = 17/58 (29%), Positives = 26/58 (44%), Gaps = 10/58 (17%)
Frame = +2
Query: 51 SYGIDFNVSREQVMKLVGRGGGCSGFSCKDIR----------FTFDAKNVLDFSLKHA 98
S+ +DF V + + ++ G SG C DIR F+ + K V +KHA
Sbjct: 266 SHAVDFTVGKFGTLHIIVNNAGISGSPCSDIRNADLSEFDKVFSVNTKGVF-HGMKHA 436
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,425,741
Number of Sequences: 63676
Number of extensions: 196174
Number of successful extensions: 961
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 951
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 955
length of query: 455
length of database: 12,639,632
effective HSP length: 100
effective length of query: 355
effective length of database: 6,272,032
effective search space: 2226571360
effective search space used: 2226571360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146751.5


