

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146747.9 - phase: 2 /pseudo
(441 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
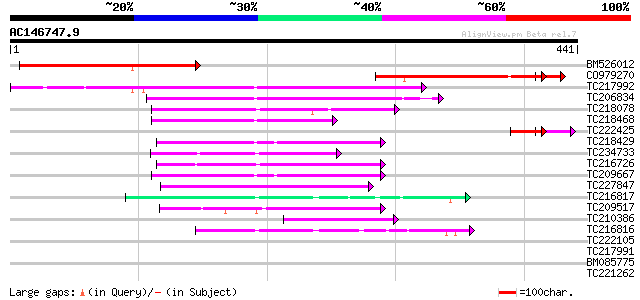
Score E
Sequences producing significant alignments: (bits) Value
BM526012 202 3e-52
CO979270 183 3e-48
TC217992 180 1e-45
TC206834 similar to UP|Q944I3 (Q944I3) AT3g01400/T13O15_4, parti... 72 6e-13
TC218078 similar to UP|Q71VM2 (Q71VM2) Plakoglobin/armadillo/bet... 68 9e-12
TC218468 67 1e-11
TC222425 weakly similar to UP|Q9FI58 (Q9FI58) Arabidopsis thalia... 49 3e-10
TC218429 similar to GB|BAC22265.1|24414015|AP003803 arm repeat c... 60 2e-09
TC234733 similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing pro... 54 1e-07
TC216726 similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing pro... 53 2e-07
TC209667 similar to GB|BAC22265.1|24414015|AP003803 arm repeat c... 53 3e-07
TC227847 similar to UP|O22161 (O22161) F-box protein family, AtF... 52 4e-07
TC216817 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like pro... 43 2e-04
TC209517 weakly similar to GB|AAP21292.1|30102748|BT006484 At1g6... 43 3e-04
TC210386 similar to UP|Q84TU4 (Q84TU4) Arm repeat-containing pro... 42 4e-04
TC216816 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like pro... 42 6e-04
TC222105 41 0.001
TC217991 41 0.001
BM085775 40 0.002
TC221262 weakly similar to UP|Q93XR9 (Q93XR9) Bg55 protein, part... 36 0.030
>BM526012
Length = 433
Score = 202 bits (513), Expect = 3e-52
Identities = 103/144 (71%), Positives = 124/144 (85%), Gaps = 3/144 (2%)
Frame = +1
Query: 8 LTQLQTHLTDFSTEYSKSSTTNPLSLHLLHSVLQTLNDAVSLSHHCQSQILPHGKLQTQS 67
LTQLQTHLTDFS E+ +ST+NPLSLHLLHS+ QTLNDAVSLS CQ + LP+GKL+TQS
Sbjct: 1 LTQLQTHLTDFSAEFPNASTSNPLSLHLLHSISQTLNDAVSLSKTCQPETLPNGKLKTQS 180
Query: 68 QIDSLIATLHRHVTDCDVLFRSGLLLE---TPAFSKREAVRSLTRNLIARLQIGSPESRA 124
+DSL+ +L RHV+DCD+LFRSGLLLE + + SKREA+RS +R+LI RLQIGSPES+A
Sbjct: 181 DLDSLLVSLDRHVSDCDILFRSGLLLENSVSVSVSKREAIRSESRSLITRLQIGSPESKA 360
Query: 125 TAIDSLLSLLNQDDKNVTIAVAQG 148
+A+DSLL LL +DDKNVTIAVAQG
Sbjct: 361 SAMDSLLGLLQEDDKNVTIAVAQG 432
>CO979270
Length = 837
Score = 183 bits (464), Expect(2) = 3e-48
Identities = 95/141 (67%), Positives = 111/141 (78%), Gaps = 8/141 (5%)
Frame = -1
Query: 285 TGLARENAIGCVANLISEDES--------MRVLAVKEGGVECLKNYWDSVTMIQSLEVGV 336
T +ARENA+GC++NL + S +RV+ VKEGGVECLKNYWDS IQSLEV V
Sbjct: 837 TAVARENAVGCLSNLTNSGSSEEADGLLNLRVMVVKEGGVECLKNYWDSGNQIQSLEVAV 658
Query: 337 EMLRYLAMTGPIDEVLVGEGFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTRKEMG 396
EMLR+LA + PI EVLVGEGFV R++GVL+C+VL VRI AARAVYA+GLN G + RKEMG
Sbjct: 657 EMLRHLAESDPIGEVLVGEGFVQRLVGVLNCEVLAVRIVAARAVYALGLNSG-RARKEMG 481
Query: 397 ECGCVPSLIKMLDGKGVEEKD 417
E GCV LIKMLDGKGVEEK+
Sbjct: 480 ELGCVLGLIKMLDGKGVEEKE 418
Score = 27.3 bits (59), Expect(2) = 3e-48
Identities = 11/23 (47%), Positives = 14/23 (60%)
Frame = -2
Query: 410 GKGVEEKDLLQWRCRCCCSTLSI 432
GK + K LL+W C+CC T I
Sbjct: 440 GKVWKRKKLLRWHCQCC*CTRRI 372
>TC217992
Length = 1097
Score = 180 bits (456), Expect = 1e-45
Identities = 118/334 (35%), Positives = 193/334 (57%), Gaps = 10/334 (2%)
Frame = +1
Query: 1 WSIFGVKLTQLQTHLTDFSTEYSKSSTTNPLSLHLLHSVLQTLNDAVSLSHHCQSQILPH 60
W + KL Q+ + L+D S+ S N L L +V +TL +A+ L+ C +
Sbjct: 55 WKMIVAKLEQIPSQLSDLSSHPCFSK--NALCKEQLQAVSKTLGEAIELAELCVKEKY-E 225
Query: 61 GKLQTQSQIDSLIATLHRHVTDCDVLFRSGLLLE-----TPAFSKRE---AVRSLTRNLI 112
GKL+ QS +D+L L ++ DC +L ++G+L E T + S E A + R L+
Sbjct: 226 GKLRMQSDLDALAGKLDLNLRDCGLLIKTGVLGEATLPLTVSSSVAESDVATHNNIRELL 405
Query: 113 ARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSD-MKEKTVAAI 171
ARLQIG E++ A+DS++ + +D+K+V + + + LV+LL ++S ++EKTV I
Sbjct: 406 ARLQIGHLEAKHRALDSVVEAMKEDEKSVLAVLGRSNIAALVQLLTATSPRIREKTVTVI 585
Query: 172 SRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRG 231
++ S +N L++EG+L L+R+++SGS + EKA I+LQ LS+S + ARAI G
Sbjct: 586 CSLAETGSCENWLVSEGVL--PPLIRLVESGSVVGKEKATISLQRLSMSAETARAIVGHG 759
Query: 232 GISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGL-ARE 290
G+ L+ +CQ G SQ AA L+N++ E+++ EE V V++ L + G L ++E
Sbjct: 760 GVRPLVALCQTGDSVSQAAAACTLKNISAVPEVRQALAEEGIVTVMINLLNCGILLGSKE 939
Query: 291 NAIGCVANLISEDESMRVLAVKEGGVECLKNYWD 324
+A C+ NL + +E++R + EGGV L Y D
Sbjct: 940 HAAECLQNLTASNENLRKSVISEGGVRSLLAYLD 1041
>TC206834 similar to UP|Q944I3 (Q944I3) AT3g01400/T13O15_4, partial (89%)
Length = 1740
Score = 71.6 bits (174), Expect = 6e-13
Identities = 64/232 (27%), Positives = 108/232 (45%), Gaps = 1/232 (0%)
Frame = +3
Query: 107 LTRNLIARLQIGS-PESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKE 165
L R L+A L S + + A++ L N+ + + IA A + P++ + ++E
Sbjct: 228 LIRQLVADLHSSSIDDQKQAAMEIRLLAKNKPENRIKIAKAGAIKPLISLISSPDLQLQE 407
Query: 166 KTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNAR 225
V AI +S + K + + G + LVR L+SG+ A E A AL LS +N
Sbjct: 408 YGVTAILNLSLCDENKEVIASSGAI--KPLVRALNSGTATAKENAACALLRLSQVEENKA 581
Query: 226 AIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGT 285
AIG G I L+ + + G ++ A+ L +L E K V+ + VL+ L +
Sbjct: 582 AIGRSGAIPLLVSLLESGGFRAKKDASTALYSLCTVKENKIRAVKAGIMKVLVELMADFE 761
Query: 286 GLARENAIGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMIQSLEVGVE 337
+ + V+ L++ E+ RV V+EGGV L ++ +EVG +
Sbjct: 762 SNMVDKSAYVVSVLVAVPEA-RVALVEEGGVPVL---------VEIVEVGTQ 887
>TC218078 similar to UP|Q71VM2 (Q71VM2)
Plakoglobin/armadillo/beta-catenin-like protein
(Fragment), partial (88%)
Length = 1310
Score = 67.8 bits (164), Expect = 9e-12
Identities = 55/198 (27%), Positives = 102/198 (50%), Gaps = 5/198 (2%)
Frame = +3
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKN-VTIAVAQGVVPVLVRLLDSSSDMKEKTVA 169
L++ L++ SPES A+ +LL+L +D+KN + I A + P++ L + +++E A
Sbjct: 294 LVSMLRVDSPESHEPALLALLNLAVKDEKNKINIVEAGALEPIISFLKSQNLNLQESATA 473
Query: 170 AISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGS 229
++ +S + K + A G++ L LV++L GS A A +AL LS +N I
Sbjct: 474 SLLTLSASSTNKPIISACGVIPL--LVQILRDGSHQAKADAVMALSNLSTHTNNLSIILE 647
Query: 230 RGGIS---SLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFV-EENAVIVLLGLASSGT 285
I LL C+ + ++ A++ +L ++E + EE V+ ++ + SGT
Sbjct: 648 TNPIPYMVDLLKTCKKSSKTAE-KCCALIESLVDYDEGRTALTSEEGGVLAVVEVLESGT 824
Query: 286 GLARENAIGCVANLISED 303
+RE+A+G + + D
Sbjct: 825 LQSREHAVGALLTMCQSD 878
>TC218468
Length = 992
Score = 67.4 bits (163), Expect = 1e-11
Identities = 47/146 (32%), Positives = 75/146 (51%), Gaps = 1/146 (0%)
Frame = +2
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDM-KEKTVA 169
L+ +L S E R A+ L SL + N + G +PVLV LL S + ++ V
Sbjct: 557 LVRKLSCRSVEERRAAVTELRSLSKRSTDNRILIAEAGAIPVLVNLLTSEDVLTQDNAVT 736
Query: 170 AISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGS 229
+I +S E+ K ++ G + +V+VL +G+ A E A L +LSL+ +N IG+
Sbjct: 737 SILNLSIYENNKGLIMLAGAI--PSIVQVLRAGTMEARENAAATLFSLSLADENKIIIGA 910
Query: 230 RGGISSLLGICQGGTPGSQGYAAAVL 255
G I +L+ + Q G+P + AA L
Sbjct: 911 SGAIPALVELLQNGSPRGKKDAATAL 988
>TC222425 weakly similar to UP|Q9FI58 (Q9FI58) Arabidopsis thaliana genomic
DNA, chromosome 5, TAC clone:K3K7, partial (23%)
Length = 410
Score = 48.5 bits (114), Expect(2) = 3e-10
Identities = 22/28 (78%), Positives = 24/28 (85%)
Frame = +2
Query: 390 KTRKEMGECGCVPSLIKMLDGKGVEEKD 417
+ RKEMGE GCV LIKMLDGKGVEEK+
Sbjct: 11 RARKEMGELGCVLGLIKMLDGKGVEEKE 94
Score = 34.3 bits (77), Expect(2) = 3e-10
Identities = 15/31 (48%), Positives = 18/31 (57%)
Frame = +3
Query: 410 GKGVEEKDLLQWRCRCCCSTLSIGGSLERMR 440
G+ +K LLQW CRCC T IGG R +
Sbjct: 72 GRVWRKKKLLQWHCRCC*CTRRIGGFSARTK 164
>TC218429 similar to GB|BAC22265.1|24414015|AP003803 arm repeat containing
protein homolog-like {Oryza sativa (japonica
cultivar-group);} , partial (39%)
Length = 985
Score = 60.1 bits (144), Expect = 2e-09
Identities = 48/180 (26%), Positives = 90/180 (49%), Gaps = 2/180 (1%)
Frame = +3
Query: 115 LQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAAISRV 174
L+ G+ S+ A +LLSL ++ +I + + P++ LL+ SS K+ + + ++
Sbjct: 9 LKTGTETSKQNAACALLSLALVEENKGSIGASGAIPPLVSLLLNGSSRGKKDALTTLYKL 188
Query: 175 STVESGKNNLLAEGLLLLNHLVR-VLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGI 233
+V K ++ G + LV V + GSG+A EKA + L +L+ ++ AI GGI
Sbjct: 189 CSVRQNKERAVSAGAV--KPLVELVAEQGSGMA-EKAMVVLNSLAGIQEGKNAIVEEGGI 359
Query: 234 SSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENF-VEENAVIVLLGLASSGTGLARENA 292
++L+ + G+ + +A L L + F V E + L+ L+ +G+ A+ A
Sbjct: 360 AALVEAIEDGSVKGKEFAVLTLLQLCVDSVRNRGFLVREGGIPPLVALSQTGSVRAKHKA 539
>TC234733 similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing protein
homolog, partial (22%)
Length = 500
Score = 53.9 bits (128), Expect = 1e-07
Identities = 43/150 (28%), Positives = 75/150 (49%), Gaps = 1/150 (0%)
Frame = +1
Query: 110 NLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVA 169
+++ L+ GS E+R A +L SL D+ V I A G +P L++LL + +K A
Sbjct: 31 DIVDVLKNGSMEARENAAATLFSLSVLDENKVQIGAA-GAIPALIKLLCEGTPRGKKDAA 207
Query: 170 -AISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIG 228
AI +S + K + G++ L++ L G +++A + L+ + AIG
Sbjct: 208 TAIFNLSIYQGNKARAVKAGIVA--PLIQFLKDAGGGMVDEALAIMAILASHHEGRVAIG 381
Query: 229 SRGGISSLLGICQGGTPGSQGYAAAVLRNL 258
I L+ + + G+P ++ AAAVL +L
Sbjct: 382 QAEPIHILVEVIRTGSPRNRENAAAVLWSL 471
>TC216726 similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing protein
homolog, partial (35%)
Length = 1115
Score = 53.1 bits (126), Expect = 2e-07
Identities = 51/180 (28%), Positives = 85/180 (46%), Gaps = 2/180 (1%)
Frame = +3
Query: 115 LQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVA-AISR 173
L+ GS E+R A +L SL D+ VTI + G +P LV LL + +K A A+
Sbjct: 117 LKKGSMEARENAAATLFSLSVIDENKVTIG-SLGAIPPLVTLLSEGNQRGKKDAATALFN 293
Query: 174 VSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGI 233
+ + K + G++ L+R+L SG +++A L L+ + I + +
Sbjct: 294 LCIYQGNKGKAVRAGVIPT--LMRLLTEPSGGMVDEALAILAILASHPEGKATIRASEAV 467
Query: 234 SSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVI-VLLGLASSGTGLARENA 292
L+ G+P ++ AAAVL +L ++ +E V+ LL LA +GT + A
Sbjct: 468 PVLVEFIGNGSPRNKENAAAVLVHLCSGDQQYLAQAQELGVMGPLLELAQNGTDRGKRKA 647
>TC209667 similar to GB|BAC22265.1|24414015|AP003803 arm repeat containing
protein homolog-like {Oryza sativa (japonica
cultivar-group);} , partial (41%)
Length = 787
Score = 52.8 bits (125), Expect = 3e-07
Identities = 47/184 (25%), Positives = 85/184 (45%), Gaps = 2/184 (1%)
Frame = +2
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAA 170
LI L+ G+ S+ A +L+SL + +I + P++ LL S K+ +
Sbjct: 47 LIYVLKTGTETSKQNAACALMSLALVKENKSSIGACGAIPPLVALLLSGSQRGKKDALTT 226
Query: 171 ISRVSTVESGKNNLLAEGLLLLNHLVR-VLDSGSGLAIEKACIALQALSLSRDNARAIGS 229
+ ++ +V K ++ G + LV V + GSG+A EKA + L +L+ + AI
Sbjct: 227 LYKLCSVRQNKERAVSAGAV--RPLVELVAEEGSGMA-EKAMVVLNSLAGIEEGKEAIVE 397
Query: 230 RGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEI-KENFVEENAVIVLLGLASSGTGLA 288
GGI +LL + G+ + +A L L + + V E + L+ L+ + + A
Sbjct: 398 EGGIGALLEAIEDGSVKGKEFAVLTLVQLCAHSVANRALLVREGGIPPLVALSQNTSVRA 577
Query: 289 RENA 292
+ A
Sbjct: 578 KLKA 589
>TC227847 similar to UP|O22161 (O22161) F-box protein family, AtFBX5, partial
(43%)
Length = 1566
Score = 52.4 bits (124), Expect = 4e-07
Identities = 46/168 (27%), Positives = 76/168 (44%), Gaps = 2/168 (1%)
Frame = +2
Query: 118 GSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSD-MKEKTVAAISRVST 176
G E A A+ +L + + + N + G + LV+L S + ++++ A+ +S
Sbjct: 185 GVQEQAARALANLAAHGDSNSNNAAVGQEAGALEALVQLTCSPHEGVRQEAAGALWNLSF 364
Query: 177 VESGKNNLLAEG-LLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGISS 235
+ + + A G + L L + + S E+A AL LS+S N+ AIG GG++
Sbjct: 365 DDRNREAIAAAGGVQALVALAQACANASPGLQERAAGALWGLSVSETNSVAIGREGGVAP 544
Query: 236 LLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASS 283
L+ + + AA L NLA VEE V L+ L SS
Sbjct: 545 LIALARSEAEDVHETAAGALWNLAFNASNALRIVEEGGVSALVDLCSS 688
>TC216817 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like protein X,
partial (5%)
Length = 1071
Score = 43.1 bits (100), Expect = 2e-04
Identities = 64/280 (22%), Positives = 112/280 (39%), Gaps = 12/280 (4%)
Frame = +1
Query: 91 LLLETPAFSKREAVRSLTRNLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVV 150
L+ +P K + + +I L ES+ A L D V +G V
Sbjct: 79 LVHSSPNIKKEVLLAGALQPVIGLLSSCCSESQREAALLLGQFAATDSDCKVHIVQRGAV 258
Query: 151 PVLVRLLDSSS-DMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEK 209
L+ +L S ++E + A+ R++ + + G L+ L+++LDS +G
Sbjct: 259 QPLIEMLQSPDVQLREMSAFALGRLAQDPHNQAGIAHNGGLV--PLLKLLDSKNGSLQHN 432
Query: 210 ACIALQALSLSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFV 269
A AL L+ + DNA GG+ L Q G Q V + L + E K +
Sbjct: 433 AAFALYGLADNEDNASDFIRVGGVQRL----QDGEFIVQATKDCVAKTLKRLEE-KIHGR 597
Query: 270 EENAVIVLLGLASSGTGLARENAIGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMI 329
N ++ L+ ++ G + +A+L S D+ R++ + G+E L S +
Sbjct: 598 VLNHLLYLMRVSEKG---CQRRVALALAHLCSSDD-QRIIFIDHYGLELLIGLLGSSSSK 765
Query: 330 QSLEVGVEMLRY-----------LAMTGPIDEVLVGEGFV 358
Q L+ V + + A P +V +GE +V
Sbjct: 766 QQLDGAVALCKLADKASTLSPVDAAPPSPTPQVYLGEQYV 885
>TC209517 weakly similar to GB|AAP21292.1|30102748|BT006484 At1g67530
{Arabidopsis thaliana;} , partial (34%)
Length = 1350
Score = 42.7 bits (99), Expect = 3e-04
Identities = 49/182 (26%), Positives = 86/182 (46%), Gaps = 6/182 (3%)
Frame = +1
Query: 117 IGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEK--TVAAISRV 174
I S A+ L+L D I +Q V L+++L++ ++++ K ++ A+ +
Sbjct: 307 ISKTSSYGCAVALYLNLSCLDKAKHMIGTSQAV-QFLIQILEAKTEVQCKIDSLHALYNL 483
Query: 175 STVESGKNNLLAEGLL--LLNHLVRVLDSGSGLAIEKACIALQALSL-SRDNARAIGSRG 231
STV S NLL+ G++ L + LV D G + EK L L++ + + + G
Sbjct: 484 STVPSNIPNLLSSGIMDGLQSLLV---DQGDCMWTEKCIAVLINLAVYQAGREKMMLAPG 654
Query: 232 GISSLLGICQGGTPGSQGYAAAVLRNLA-KFNEIKENFVEENAVIVLLGLASSGTGLARE 290
IS+L G P Q AA+ L L + E + ++E + L+ ++ +GT RE
Sbjct: 655 LISALASTLDTGEPIEQEQAASCLLILCNRSEECCQMVLQEGVIPALVSISVNGTSRGRE 834
Query: 291 NA 292
A
Sbjct: 835 KA 840
Score = 29.6 bits (65), Expect = 2.8
Identities = 17/57 (29%), Positives = 31/57 (53%), Gaps = 1/57 (1%)
Frame = +1
Query: 111 LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRL-LDSSSDMKEK 166
L + L G P + A LL L N+ ++ + + +GV+P LV + ++ +S +EK
Sbjct: 667 LASTLDTGEPIEQEQAASCLLILCNRSEECCQMVLQEGVIPALVSISVNGTSRGREK 837
>TC210386 similar to UP|Q84TU4 (Q84TU4) Arm repeat-containing protein,
partial (13%)
Length = 653
Score = 42.4 bits (98), Expect = 4e-04
Identities = 30/90 (33%), Positives = 46/90 (50%), Gaps = 1/90 (1%)
Frame = +1
Query: 214 LQALSLSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKENFV-EEN 272
L L+ + AIG +GGI L+ + + G+ + AAA L +L N N V +E
Sbjct: 7 LANLATIPEGKTAIGQQGGIPVLVEVIESGSARGKENAAAALLHLCSDNHRYLNMVLQEG 186
Query: 273 AVIVLLGLASSGTGLARENAIGCVANLISE 302
AV L+ L+ SGT A+E A+ + S+
Sbjct: 187 AVPPLVALSQSGTPRAKEKALALLNQFRSQ 276
Score = 35.8 bits (81), Expect = 0.039
Identities = 20/64 (31%), Positives = 34/64 (52%)
Frame = +1
Query: 253 AVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLARENAIGCVANLISEDESMRVLAVK 312
AVL NLA E K ++ + VL+ + SG+ +ENA + +L S++ + ++
Sbjct: 1 AVLANLATIPEGKTAIGQQGGIPVLVEVIESGSARGKENAAAALLHLCSDNHRYLNMVLQ 180
Query: 313 EGGV 316
EG V
Sbjct: 181 EGAV 192
>TC216816 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like protein X,
partial (5%)
Length = 1610
Score = 42.0 bits (97), Expect = 6e-04
Identities = 60/229 (26%), Positives = 95/229 (41%), Gaps = 12/229 (5%)
Frame = +1
Query: 145 VAQGVVPVLVRLLDSSS-DMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGS 203
V +G V L+ +L SS +KE + A+ R++ + + G L+ L+++LDS +
Sbjct: 61 VQRGAVRPLIEMLQSSDVQLKEMSAFALGRLAQDTHNQAGIAHNGGLM--PLLKLLDSKN 234
Query: 204 GLAIEKACIALQALSLSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNE 263
G A AL L+ + DN GGI L Q G Q V + L + E
Sbjct: 235 GSLQHNAAFALYGLADNEDNVSDFIRVGGIQRL----QDGEFIVQATKDCVAKTLKRLEE 402
Query: 264 IKENFVEENAVIVLLGLASSGTGLARENAIGCVANLISEDESMRVLAVKEGGVECLKNYW 323
V + +L + S R A+ +A+L S D+ ++ + G+E L
Sbjct: 403 KIHGRVLNH---LLYLMRVSEKAFQRRVAL-TLAHLCSADDQRKIF-IDYNGLELLMGLL 567
Query: 324 DSVTMIQSLEVGVEM--LRYLAMT---------GPIDEVLVGEGFVGRV 361
S Q L+ V + L AMT P +V +GE +V V
Sbjct: 568 GSYNPKQQLDGAVALCKLANKAMTLSPVDAAPPSPTPQVYLGEQYVNNV 714
>TC222105
Length = 904
Score = 41.2 bits (95), Expect = 0.001
Identities = 47/187 (25%), Positives = 80/187 (42%), Gaps = 4/187 (2%)
Frame = +1
Query: 145 VAQGVVPVLVRLLDSSSDMKEKTVAAISRVSTVESGKNNLLAEGLL-LLNHLVRVLDSGS 203
V G VP L+ +L + E A + ++ +N +A G+L L+ L+ L + S
Sbjct: 28 VRSGFVPFLIDVLKGG--LGESQEHAAGALFSLALDDDNKMAIGVLGALHPLMHALRAES 201
Query: 204 GLAIEKACIALQALSLSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNE 263
+ +AL LSL + N + G + +LL + G S+ +L NLA E
Sbjct: 202 ERTRHDSALALYHLSLVQSNRMKLVKLGVVPTLLSMVVAGNLASR--VLLILCNLAVCTE 375
Query: 264 IKENFVEENAVIVLLGLASSG---TGLARENAIGCVANLISEDESMRVLAVKEGGVECLK 320
+ ++ NAV +L+ L + REN + + L + LA + E LK
Sbjct: 376 GRTAMLDANAVEILVSLLRGNELDSEATRENCVAALYALSHRSLRFKGLAKEARVAEVLK 555
Query: 321 NYWDSVT 327
++ T
Sbjct: 556 EIEETGT 576
>TC217991
Length = 694
Score = 40.8 bits (94), Expect = 0.001
Identities = 29/79 (36%), Positives = 41/79 (51%)
Frame = +3
Query: 333 EVGVEMLRYLAMTGPIDEVLVGEGFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTR 392
E V LR L + P +E LV G + R+ VL L + AAA A+ + +
Sbjct: 12 ESAVGALRNLVGSVP-EESLVSLGLIPRLAHVLKSGSLGAQQAAAAAICRVC--SSTDMK 182
Query: 393 KEMGECGCVPSLIKMLDGK 411
K +GE GC+P L+KML+ K
Sbjct: 183 KMVGEAGCIPLLVKMLEAK 239
>BM085775
Length = 422
Score = 40.0 bits (92), Expect = 0.002
Identities = 35/119 (29%), Positives = 56/119 (46%), Gaps = 1/119 (0%)
Frame = +2
Query: 182 NNLLAEGLL-LLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGISSLLGIC 240
+N +A G+L L+ L+ L + S + +AL LSL + N + G + +LL +
Sbjct: 38 DNKMAIGVLGALHPLMHALRAESERTRHDSALALYHLSLVQSNRLKLVKLGAVPTLLSMV 217
Query: 241 QGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLARENAIGCVANL 299
G S+ +L NLA E + ++ NAV +L+GL + N CVA L
Sbjct: 218 VAGNLASR--VLLILCNLAVCTEGRTAMLDANAVEILVGLLRGNELDSEANRENCVAAL 388
>TC221262 weakly similar to UP|Q93XR9 (Q93XR9) Bg55 protein, partial (10%)
Length = 935
Score = 36.2 bits (82), Expect = 0.030
Identities = 19/65 (29%), Positives = 36/65 (55%)
Frame = +1
Query: 110 NLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVA 169
+++ L+ GS E + A+ LLSL +Q + + V +G++P LV + + SDM +
Sbjct: 91 SVVEILETGSDEEKEPALIILLSLCSQRVEYCQLVVYEGIIPSLVNISNKGSDMAKAYAL 270
Query: 170 AISRV 174
+ R+
Sbjct: 271 ELLRL 285
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,797,546
Number of Sequences: 63676
Number of extensions: 240884
Number of successful extensions: 1821
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 1787
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1808
length of query: 441
length of database: 12,639,632
effective HSP length: 100
effective length of query: 341
effective length of database: 6,272,032
effective search space: 2138762912
effective search space used: 2138762912
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146747.9


