

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.7 + phase: 0
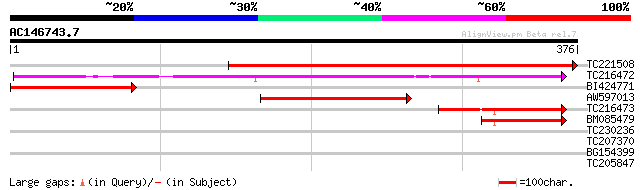
(376 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221508 similar to UP|Q8LF92 (Q8LF92) Homoserine dehydrogenase-... 411 e-115
TC216472 UP|O63067 (O63067) Aspartokinase-homoserine dehydrogena... 188 4e-48
BI424771 weakly similar to GP|21537197|gb| homoserine dehydrogen... 127 8e-30
AW597013 similar to PIR|T06246|T06 aspartate kinase (EC 2.7.2.4)... 80 1e-15
TC216473 homologue to UP|O63067 (O63067) Aspartokinase-homoserin... 65 5e-11
BM085479 homologue to PIR|T06242|T062 aspartate kinase (EC 2.7.2... 52 4e-07
TC230236 similar to UP|Q940D2 (Q940D2) SKIP5-like protein, parti... 30 1.4
TC207370 similar to UP|Q93ZI7 (Q93ZI7) AT5g35440/MOK9_2, partial... 29 4.0
BG154399 28 8.8
TC205847 homologue to UP|Q6X7J9 (Q6X7J9) WOX4 protein (At1g46480... 28 8.8
>TC221508 similar to UP|Q8LF92 (Q8LF92) Homoserine dehydrogenase-like
protein, partial (60%)
Length = 912
Score = 411 bits (1056), Expect = e-115
Identities = 209/231 (90%), Positives = 219/231 (94%)
Frame = +2
Query: 146 TMGDFEKLLTYPRRIRHESTVGAGLPVISSLNRIISSGDPVNRIIGSLSGTLGYVMSEVE 205
TM DF+KL YPRRIRHESTVGAGLPVI+SLNRIISSGDPV++IIGSLSGTLGYVMSEVE
Sbjct: 2 TMEDFKKLFIYPRRIRHESTVGAGLPVIASLNRIISSGDPVHQIIGSLSGTLGYVMSEVE 181
Query: 206 DGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVARKALILARILGRRINMDSIQIESLYPKE 265
DGKPLSQVVR AKSLGYTEPDPRDDLGGMDVARKALILARILG +IN+DSIQIESLYPKE
Sbjct: 182 DGKPLSQVVRDAKSLGYTEPDPRDDLGGMDVARKALILARILGHQINLDSIQIESLYPKE 361
Query: 266 MGPDVMTDDDFLSCGLLLLDKDIQERVEKAASNGNVLRYVCVIEGPRCEVGIQELPKNSA 325
MGP VMT +DFL+ GLLLLDKDIQ+RVEKAASNGNVLRYVCVI G RCEVGIQELPKNS
Sbjct: 362 MGPGVMTVEDFLNRGLLLLDKDIQDRVEKAASNGNVLRYVCVIMGSRCEVGIQELPKNSP 541
Query: 326 LGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGNDTTAAGVLADIVDIQDLFP 376
LGRLRGSDNVLE+YTRCYS QPLVIQGAGAGNDTTAAGVLADIVDIQDLFP
Sbjct: 542 LGRLRGSDNVLEIYTRCYSKQPLVIQGAGAGNDTTAAGVLADIVDIQDLFP 694
>TC216472 UP|O63067 (O63067) Aspartokinase-homoserine dehydrogenase ,
complete
Length = 3148
Score = 188 bits (477), Expect = 4e-48
Identities = 129/374 (34%), Positives = 196/374 (51%), Gaps = 7/374 (1%)
Frame = +2
Query: 3 TIPLILMGCGGVGSHLLQHIVSSRS-LHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
TI + ++G G +GS LL + S L + LRV+GI SKS+++ D G D
Sbjct: 1697 TIAMGIIGPGLIGSTLLDQLRDQASTLKEEFNIDLRVMGILGSKSMLLSD----VGID-- 1858
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L E + G++ + FV + A VDCTA S
Sbjct: 1859 -------LARWRELREERGEVANMEKFVQHVHGNHFIP---------NTALVDCTADSAI 1990
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIR----HESTVGAGLPVISSLN 177
+ G VV NKK + + + KL R+ +E+TVGAGLP++S+L
Sbjct: 1991 AGYYYDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLR 2170
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++ +GD + +I G SGTL Y+ + +DG+ S+VV AK GYTEPDPRDDL G DVA
Sbjct: 2171 GLLETGDKILQIEGIFSGTLSYIFNNFKDGRAFSEVVSEAKEAGYTEPDPRDDLSGTDVA 2350
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK +ILAR G ++ + +I +ESL P+ + + +F+ L D++ ++ E A +
Sbjct: 2351 RKVIILARESGLKLELSNIPVESLVPEPLRA-CASAQEFMQ-ELPKFDQEFTKKQEDAEN 2524
Query: 298 NGNVLRYVCVIE--GPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV V++ + V ++ K+ +L GSDN++ TR Y +QPL+++G GA
Sbjct: 2525 AGEVLRYVGVVDVTNKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGA 2704
Query: 356 GNDTTAAGVLADIV 369
G TA G+ +DI+
Sbjct: 2705 GAQVTAGGIFSDIL 2746
>BI424771 weakly similar to GP|21537197|gb| homoserine dehydrogenase-like
protein {Arabidopsis thaliana}, partial (21%)
Length = 304
Score = 127 bits (319), Expect = 8e-30
Identities = 64/84 (76%), Positives = 71/84 (84%)
Frame = +2
Query: 1 MKTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDD 60
MK IPLILMGCGGVG LLQHIVS RSLHS+QGLCLRVVG+GDSKSLVV D LL++G +D
Sbjct: 53 MKNIPLILMGCGGVGRQLLQHIVSCRSLHSTQGLCLRVVGVGDSKSLVVTDYLLHEGLND 232
Query: 61 SFLLELCRLKHGGESLSKLGDLGQ 84
FLLELCR+K GESL KL D G+
Sbjct: 233 GFLLELCRVKSVGESLLKLLDFGK 304
>AW597013 similar to PIR|T06246|T06 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean (fragment), partial (12%)
Length = 334
Score = 80.5 bits (197), Expect = 1e-15
Identities = 38/100 (38%), Positives = 63/100 (63%)
Frame = +2
Query: 167 GAGLPVISSLNRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPD 226
G GLP++S+L ++ +GD + +I G GTL Y+ + +DG+ S+ +K YT+P+
Sbjct: 5 GPGLPIVSTLRGLLETGDKILQIEGIFIGTLSYIFNNFKDGRAYSEESSESKEARYTDPN 184
Query: 227 PRDDLGGMDVARKALILARILGRRINMDSIQIESLYPKEM 266
P DDL G D A KA+ILA+ G ++ + +I +SL P+ +
Sbjct: 185 PIDDLSGTDAAIKAIILAKESGLKLELTNIPDDSLEPEPL 304
>TC216473 homologue to UP|O63067 (O63067) Aspartokinase-homoserine
dehydrogenase , partial (11%)
Length = 604
Score = 65.1 bits (157), Expect = 5e-11
Identities = 35/88 (39%), Positives = 54/88 (60%), Gaps = 3/88 (3%)
Frame = +2
Query: 285 DKDIQERVEKAASNGNVLRYVCVIEGPRCEVGIQEL---PKNSALGRLRGSDNVLEVYTR 341
D++ ++ E A + G VLRYV V++ E G+ EL K+ +L GSDN++ TR
Sbjct: 5 DQEFTKKQEDAENAGEVLRYVGVVDVTN-EKGVVELRRYKKDHPFAQLSGSDNIIAFTTR 181
Query: 342 CYSNQPLVIQGAGAGNDTTAAGVLADIV 369
Y +QPL+++G GAG TA G+ +DI+
Sbjct: 182 RYKDQPLIVRGPGAGAQVTAGGIFSDIL 265
>BM085479 homologue to PIR|T06242|T062 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (8%)
Length = 347
Score = 52.0 bits (123), Expect = 4e-07
Identities = 26/59 (44%), Positives = 36/59 (60%), Gaps = 3/59 (5%)
Frame = +1
Query: 314 EVGIQEL---PKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGNDTTAAGVLADIV 369
E G+ EL K+ +L GSDN++ TR Y NQPL+++G GAG TA G+ DI+
Sbjct: 19 EKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYRNQPLIVRGPGAGAQVTAGGIFNDIL 195
>TC230236 similar to UP|Q940D2 (Q940D2) SKIP5-like protein, partial (70%)
Length = 810
Score = 30.4 bits (67), Expect = 1.4
Identities = 14/35 (40%), Positives = 20/35 (57%)
Frame = +1
Query: 308 IEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRC 342
I P C +G E+P ++ L RGSD+ LE + C
Sbjct: 199 IRKPLCLIGAGEVPDDTTLVCSRGSDSALEFLSTC 303
>TC207370 similar to UP|Q93ZI7 (Q93ZI7) AT5g35440/MOK9_2, partial (15%)
Length = 1647
Score = 28.9 bits (63), Expect = 4.0
Identities = 21/56 (37%), Positives = 27/56 (47%)
Frame = +3
Query: 22 IVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDSFLLELCRLKHGGESLS 77
I SSR G+ + VVGIG + LVV D + G DS + C + G LS
Sbjct: 270 IKSSRVPSEKLGVGVCVVGIGKWRQLVVEDQISGNGLVDSSEGDDCPSEDGRLKLS 437
>BG154399
Length = 301
Score = 27.7 bits (60), Expect = 8.8
Identities = 13/20 (65%), Positives = 15/20 (75%)
Frame = -2
Query: 100 IASQLGKKTGLAFVDCTASS 119
IA GKKTGLAF+D T +S
Sbjct: 156 IAEGGGKKTGLAFIDFTCTS 97
>TC205847 homologue to UP|Q6X7J9 (Q6X7J9) WOX4 protein (At1g46480), partial
(43%)
Length = 926
Score = 27.7 bits (60), Expect = 8.8
Identities = 16/47 (34%), Positives = 27/47 (57%)
Frame = -2
Query: 175 SLNRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLG 221
SL ++SS DP+ + +LSG + + S+V G ++V +SLG
Sbjct: 241 SLLLVLSSEDPIPSFLEALSGLMKLLRSKVVTG**GGEMVSVLESLG 101
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.139 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,125,010
Number of Sequences: 63676
Number of extensions: 199939
Number of successful extensions: 799
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 796
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 798
length of query: 376
length of database: 12,639,632
effective HSP length: 99
effective length of query: 277
effective length of database: 6,335,708
effective search space: 1754991116
effective search space used: 1754991116
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146743.7


